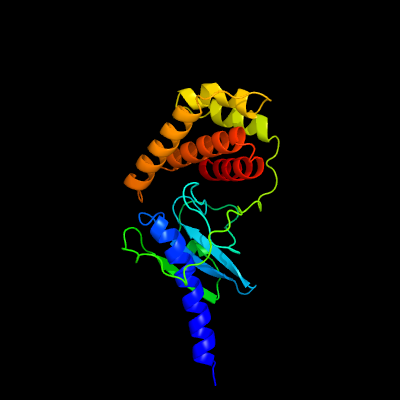
1 c5imuA_
100.0
34
PDB header: signaling proteinChain: A: PDB Molecule: tat (twin-arginine translocation) pathway signal sequencePDBTitle: a fragment of conserved hypothetical protein rv3899c (residues 184-2 410) from mycobacterium tuberculosis
2 c5ganC_
42.5
24
PDB header: transcriptionChain: C: PDB Molecule: pre-mrna-splicing factor snu114;PDBTitle: the overall structure of the yeast spliceosomal u4/u6.u5 tri-snrnp at2 3.7 angstrom
3 c3jb9B_
37.3
27
PDB header: rna binding protein/rnaChain: B: PDB Molecule: pre-mrna-splicing factor cwf10;PDBTitle: cryo-em structure of the yeast spliceosome at 3.6 angstrom resolution
4 c5mqfB_
35.5
35
PDB header: splicingChain: B: PDB Molecule: 116 kda u5 small nuclear ribonucleoprotein component;PDBTitle: cryo-em structure of a human spliceosome activated for step 2 of2 splicing (c* complex)
5 d1afra_
35.1
20
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: Ribonucleotide reductase-like
6 c6cknA_
35.0
20
PDB header: dna binding proteinChain: A: PDB Molecule: protein af-10;PDBTitle: crystal structure of an af10 fragment
7 c5lj3C_
32.1
24
PDB header: splicingChain: C: PDB Molecule: pre-mrna-splicing factor snu114;PDBTitle: structure of the core of the yeast spliceosome immediately after2 branching
8 c3en0A_
24.5
18
PDB header: hydrolaseChain: A: PDB Molecule: cyanophycinase;PDBTitle: the structure of cyanophycinase
9 d1bola_
23.8
50
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like
10 c3t0oA_
22.2
63
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease t2;PDBTitle: crystal structure analysis of human rnase t2
11 d1iooa_
21.0
63
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like
12 c5z58C_
20.3
36
PDB header: splicingChain: C: PDB Molecule: 116 kda u5 small nuclear ribonucleoprotein component;PDBTitle: cryo-em structure of a human activated spliceosome (early bact) at 4.92 angstrom.
13 c1nchA_
19.7
40
PDB header: cell adhesion proteinChain: A: PDB Molecule: n-cadherin;PDBTitle: structural basis of cell-cell adhesion by cadherins
14 d1iqqa_
19.6
24
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like
15 d2fc6a1
19.0
46
Fold: CCCH zinc fingerSuperfamily: CCCH zinc fingerFamily: CCCH zinc finger
16 d1ucda_
19.0
63
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like
17 d1sgla_
18.5
50
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like
18 d1d0gr3
17.6
60
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
19 d1jy5a_
17.5
50
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like
20 c2mv3A_
16.6
23
PDB header: nucleotide binding proteinChain: A: PDB Molecule: mitochondrial inner membrane i-aaa protease supercomplexPDBTitle: the n-domain of the aaa metalloproteinase yme1 from saccharomyces2 cerevisiae
21 d1d4va3
not modelled
16.4
60
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
22 c2fc6A_
not modelled
16.2
46
PDB header: transcriptionChain: A: PDB Molecule: target of egr1, member 1;PDBTitle: solution structure of the zf-ccch domain of target of egr1,2 member 1 (nuclear)
23 c5ancK_
not modelled
15.1
12
PDB header: translationChain: K: PDB Molecule: elongation factor tu gtp-binding domain-containing proteinPDBTitle: mechanism of eif6 release from the nascent 60s ribosomal subunit
24 d1dixa_
not modelled
15.1
31
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like
25 d2p0ta1
not modelled
14.6
28
Fold: PSPTO4464-likeSuperfamily: PSPTO4464-likeFamily: PSPTO4464-like
26 c2p0tA_
not modelled
14.6
28
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0307 protein pspto_4464;PDBTitle: structural genomics, the crystal structure of a conserved putative2 protein from pseudomonas syringae pv. tomato str. dc3000
27 c4k59A_
not modelled
14.1
36
PDB header: rna binding proteinChain: A: PDB Molecule: rna binding protein rsmf;PDBTitle: crystal structure of pseudomonas aeruginosa rsmf
28 c2pqxA_
not modelled
14.0
50
PDB header: hydrolaseChain: A: PDB Molecule: ribonuclease i;PDBTitle: e. coli rnase 1 (in vivo folded)
29 c3ff8B_
not modelled
13.5
60
PDB header: cell adhesion/immune systemChain: B: PDB Molecule: epithelial cadherin;PDBTitle: structure of nk cell receptor klrg1 bound to e-cadherin
30 d1iyba_
not modelled
13.4
29
Fold: Ribonuclease Rh-likeSuperfamily: Ribonuclease Rh-likeFamily: Ribonuclease Rh-like
31 c3d3zA_
not modelled
12.6
35
PDB header: hydrolaseChain: A: PDB Molecule: actibind;PDBTitle: crystal structure of actibind a t2 rnase
32 c1vd3A_
not modelled
12.4
29
PDB header: hydrolaseChain: A: PDB Molecule: rnase ngr3;PDBTitle: ribonuclease nt in complex with 2'-ump
33 c3jcrB_
not modelled
12.4
35
PDB header: splicingChain: B: PDB Molecule: hsnu114;PDBTitle: 3d structure determination of the human*u4/u6.u5* tri-snrnp complex
34 c2r9aA_
not modelled
12.0
31
PDB header: protein bindingChain: A: PDB Molecule: non-homologous end-joining factor 1;PDBTitle: crystal structure of human xlf
35 c6fcxA_
not modelled
11.9
41
PDB header: oxidoreductaseChain: A: PDB Molecule: methylenetetrahydrofolate reductase;PDBTitle: structure of human 5,10-methylenetetrahydrofolate reductase (mthfr)
36 d1ncia_
not modelled
11.6
43
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cadherin-likeFamily: Cadherin
37 c1nciA_
not modelled
11.6
43
PDB header: cell adhesion proteinChain: A: PDB Molecule: n-cadherin;PDBTitle: structural basis of cell-cell adhesion by cadherins
38 c5frgA_
not modelled
11.5
27
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1
39 c4bfhA_
not modelled
11.3
50
PDB header: hydrolase inhibitorChain: A: PDB Molecule: wrightide r1;PDBTitle: crystal structure of alpha-amylase inhibitor wrightide r1 (wr1)2 peptide from wrightia religiosa
40 c2mauA_
not modelled
11.0
50
PDB header: hydrolase inhibitorChain: A: PDB Molecule: wrightide r1;PDBTitle: solution structure of alpha-amylase inhibitor wrightide r1 (wr1)2 peptide from wrightia religiosa
41 d3er7a1
not modelled
10.8
58
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Exig0174-like
42 c5yrzD_
not modelled
10.3
100
PDB header: antitoxin/hydrolaseChain: D: PDB Molecule: hica;PDBTitle: toxin-antitoxin complex from streptococcus pneumoniae
43 c5yrzB_
not modelled
10.3
100
PDB header: antitoxin/hydrolaseChain: B: PDB Molecule: hica;PDBTitle: toxin-antitoxin complex from streptococcus pneumoniae
44 c6k6lA_
not modelled
10.0
50
PDB header: unknown functionChain: A: PDB Molecule: pseudo deubiquitinase;PDBTitle: ygl082w-catalytic domain
45 c1nchB_
not modelled
9.9
55
PDB header: cell adhesion proteinChain: B: PDB Molecule: n-cadherin;PDBTitle: structural basis of cell-cell adhesion by cadherins
46 c3t97A_
not modelled
9.4
33
PDB header: protein transportChain: A: PDB Molecule: nuclear pore glycoprotein p62;PDBTitle: molecular architecture of the transport channel of the nuclear pore2 complex: nup62/nup54
47 c3uwsA_
not modelled
9.4
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein;PDBTitle: crystal structure of a clostripain (parmer_00083) from parabacteroides2 merdae atcc 43184 at 1.70 a resolution
48 c5jknA_
not modelled
9.4
38
PDB header: hydrolaseChain: A: PDB Molecule: protein fam63a;PDBTitle: crystal structure of deubiquitinase mindy-1
49 c1zvnA_
not modelled
9.3
25
PDB header: cell adhesionChain: A: PDB Molecule: cadherin 1;PDBTitle: crystal structure of chick mn-cadherin ec1
50 d1gv0a2
not modelled
9.2
63
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain
51 c5gkeB_
not modelled
9.1
42
PDB header: hydrolase/dnaChain: B: PDB Molecule: endonuclease endoms;PDBTitle: structure of endoms-dsdna1 complex
52 d2omwb1
not modelled
8.9
50
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cadherin-likeFamily: Cadherin
53 c6cg7A_
not modelled
8.8
27
PDB header: cell adhesionChain: A: PDB Molecule: cadherin-22;PDBTitle: mouse cadherin-22 ec1-2 adhesive fragment
54 d1kifa1
not modelled
8.7
17
Fold: Nucleotide-binding domainSuperfamily: Nucleotide-binding domainFamily: D-aminoacid oxidase, N-terminal domain
55 d1guza2
not modelled
8.7
50
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain
56 c2axcA_
not modelled
8.6
20
PDB header: hydrolaseChain: A: PDB Molecule: colicin e7;PDBTitle: crystal structure of cole7 translocation domain
57 c2lm0A_
not modelled
8.3
13
PDB header: nuclear proteinChain: A: PDB Molecule: af4/fmr2 family member 1/protein af-9 chimera;PDBTitle: solution structure of the af4-af9 complex
58 c4ozwA_
not modelled
8.1
22
PDB header: lyaseChain: A: PDB Molecule: alginate lyase;PDBTitle: crystal structure of the periplasmic alginate lyase algl h202a mutant
59 c1q67B_
not modelled
8.0
21
PDB header: transcriptionChain: B: PDB Molecule: decapping protein involved in mrna degradation-PDBTitle: crystal structure of dcp1p
60 d1uxja2
not modelled
7.9
50
Fold: LDH C-terminal domain-likeSuperfamily: LDH C-terminal domain-likeFamily: Lactate & malate dehydrogenases, C-terminal domain
61 c2kwtA_
not modelled
7.8
67
PDB header: viral proteinChain: A: PDB Molecule: protease ns2-3;PDBTitle: solution structure of ns2 [27-59]
62 c4eo1A_
not modelled
7.6
43
PDB header: viral proteinChain: A: PDB Molecule: attachment protein g3p;PDBTitle: crystal structure of the tola binding domain from the filamentous2 phage ike
63 c2ju0B_
not modelled
7.6
41
PDB header: metal binding protein/signaling proteinChain: B: PDB Molecule: phosphatidylinositol 4-kinase pik1;PDBTitle: structure of yeast frequenin bound to pdtins 4-kinase
64 c4g4mA_
not modelled
7.5
50
PDB header: de novo proteinChain: A: PDB Molecule: alpha4f3(6-13);PDBTitle: crystal structure of the de novo designed fluorinated peptide2 alpha4f3(6-13)
65 c4g4mB_
not modelled
7.5
50
PDB header: de novo proteinChain: B: PDB Molecule: alpha4f3(6-13);PDBTitle: crystal structure of the de novo designed fluorinated peptide2 alpha4f3(6-13)
66 c4o2tB_
not modelled
7.5
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf4827 family protein (bdi_1692) from2 parabacteroides distasonis atcc 8503 at 2.40 a resolution
67 c1yzxB_
not modelled
7.3
16
PDB header: transferaseChain: B: PDB Molecule: glutathione s-transferase kappa 1;PDBTitle: crystal structure of human kappa class glutathione transferase
68 c2x3mA_
not modelled
7.3
29
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein orf239;PDBTitle: crystal structure of hypothetical protein orf239 from pyrobaculum2 spherical virus
69 c3l32B_
not modelled
7.2
14
PDB header: viral proteinChain: B: PDB Molecule: phosphoprotein;PDBTitle: structure of the dimerisation domain of the rabies virus2 phosphoprotein
70 d2cvea2
not modelled
7.2
41
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: YigZ C-terminal domain-like
71 d2cuab_
not modelled
7.2
31
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Periplasmic domain of cytochrome c oxidase subunit II
72 c1yxeA_
not modelled
7.1
57
PDB header: immune systemChain: A: PDB Molecule: apical membrane antigen 1;PDBTitle: structure and inter-domain interactions of domain ii from the blood2 stage malarial protein, apical membrane antigen 1
73 c3ejkA_
not modelled
6.9
38
PDB header: isomeraseChain: A: PDB Molecule: dtdp sugar isomerase;PDBTitle: crystal structure of dtdp sugar isomerase (yp_390184.1) from2 desulfovibrio desulfuricans g20 at 1.95 a resolution
74 c2pmpA_
not modelled
6.7
18
PDB header: lyaseChain: A: PDB Molecule: 2-c-methyl-d-erythritol 2,4-cyclodiphosphate synthase;PDBTitle: structure of 2c-methyl-d-erythritol 2,4-cyclodiphosphate synthase from2 the isoprenoid biosynthetic pathway of arabidopsis thaliana
75 c2ke4A_
not modelled
6.6
22
PDB header: membrane proteinChain: A: PDB Molecule: cdc42-interacting protein 4;PDBTitle: the nmr structure of the tc10 and cdc42 interacting domain2 of cip4
76 c5wbuR_
not modelled
6.4
63
PDB header: transferaseChain: R: PDB Molecule: proline-rich akt1 substrate 1;PDBTitle: crystal structure of mtor(deltan)-mlst8-pras40(alpha-helix & beta-2 strand) complex
77 c5wbuQ_
not modelled
6.4
63
PDB header: transferaseChain: Q: PDB Molecule: proline-rich akt1 substrate 1;PDBTitle: crystal structure of mtor(deltan)-mlst8-pras40(alpha-helix & beta-2 strand) complex
78 d1ejxb_
not modelled
6.3
44
Fold: beta-clipSuperfamily: Urease, beta-subunitFamily: Urease, beta-subunit
79 c3j9tb_
not modelled
6.2
71
PDB header: hydrolaseChain: B: PDB Molecule: v-type proton atpase subunit b;PDBTitle: yeast v-atpase state 1
80 c5fujB_
not modelled
6.2
63
PDB header: oxidoreductaseChain: B: PDB Molecule: mroupo;PDBTitle: crystallization of a dimeric heme peroxygenase from the2 fungus marasmius rotula
81 c3fybA_
not modelled
6.1
41
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: protein of unknown function (duf1244);PDBTitle: crystal structure of a protein of unknown function (duf1244) from2 alcanivorax borkumensis
82 c3biyA_
not modelled
6.1
20
PDB header: transferaseChain: A: PDB Molecule: histone acetyltransferase p300;PDBTitle: crystal structure of p300 histone acetyltransferase domain in complex2 with a bisubstrate inhibitor, lys-coa
83 c2kneB_
not modelled
6.1
67
PDB header: metal transportChain: B: PDB Molecule: atpase, ca++ transporting, plasma membrane 4;PDBTitle: calmodulin wraps around its binding domain in the plasma2 membrane ca2+ pump anchored by a novel 18-1 motif
84 c2mzyA_
not modelled
6.1
56
PDB header: iron binding proteinChain: A: PDB Molecule: probable fe(2+)-trafficking protein;PDBTitle: 1h, 13c, and 15n chemical shift assignments and structure of probable2 fe(2+)-trafficking protein from burkholderia pseudomallei 1710b.
85 c2mitA_
not modelled
6.0
86
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin-5;PDBTitle: solution structure of oxidized dimeric form of human defensin 5
86 d2omzb1
not modelled
5.9
42
Fold: Immunoglobulin-like beta-sandwichSuperfamily: Cadherin-likeFamily: Cadherin
87 c1cffB_
not modelled
5.9
67
PDB header: calmodulinChain: B: PDB Molecule: calcium pump;PDBTitle: nmr solution structure of a complex of calmodulin with a binding2 peptide of the ca2+-pump
88 d1lmb3_
not modelled
5.9
18
Fold: lambda repressor-like DNA-binding domainsSuperfamily: lambda repressor-like DNA-binding domainsFamily: Phage repressors
89 c4e86H_
not modelled
5.9
86
PDB header: antimicrobial proteinChain: H: PDB Molecule: defensin-5;PDBTitle: crystal structure of human alpha-defensin 5, hd5 (leu29aba mutant)
90 c4e86D_
not modelled
5.9
86
PDB header: antimicrobial proteinChain: D: PDB Molecule: defensin-5;PDBTitle: crystal structure of human alpha-defensin 5, hd5 (leu29aba mutant)
91 d2qtva2
not modelled
5.9
38
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: beta-sandwich domain of Sec23/24Family: beta-sandwich domain of Sec23/24
92 c4rbxA_
not modelled
5.9
86
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin-5;PDBTitle: crystal structure of human alpha-defensin 5, hd5 (glu21arg mutant)
93 d2eyqa2
not modelled
5.9
23
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain
94 c1zmpA_
not modelled
5.8
86
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin 5;PDBTitle: crystal structure of human defensin-5
95 c2lxzA_
not modelled
5.8
86
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin-5;PDBTitle: solution structure of the antimicrobial peptide human defensin 5
96 c1zmpB_
not modelled
5.8
86
PDB header: antimicrobial proteinChain: B: PDB Molecule: defensin 5;PDBTitle: crystal structure of human defensin-5
97 c4z42B_
not modelled
5.8
22
PDB header: hydrolaseChain: B: PDB Molecule: urease subunit beta;PDBTitle: crystal structure of urease from yersinia enterocolitica
98 c1zmpC_
not modelled
5.8
86
PDB header: antimicrobial proteinChain: C: PDB Molecule: defensin 5;PDBTitle: crystal structure of human defensin-5
99 c4e83A_
not modelled
5.8
86
PDB header: antimicrobial proteinChain: A: PDB Molecule: defensin-5;PDBTitle: crystal structure of human alpha-defensin 5, hd5 (leu29nle mutant)