| 1 | c2b81D_
|
|
 |
100.0 |
21 |
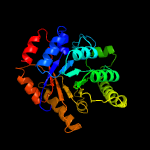
PDB header:oxidoreductase
Chain: D: PDB Molecule:luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus
|
|
|
|
| 2 | c3sdoB_
|
|
 |
100.0 |
22 |
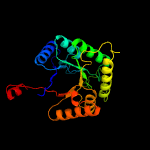
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
|
|
|
| 3 | c5tlcA_
|
|
 |
100.0 |
20 |
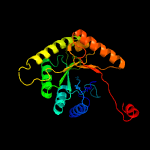
PDB header:oxidoreductase
Chain: A: PDB Molecule:dibenzothiophene desulfurization enzyme a;
PDBTitle: crystal structure of bdsa from bacillus subtilis wu-s2b
|
|
|
|
| 4 | d1tvla_
|
|
 |
100.0 |
17 |
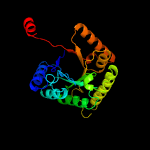
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
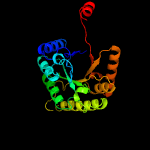
| 5 | c1tvlA_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein ytnj;
PDBTitle: structure of ytnj from bacillus subtilis
|
|
|
|
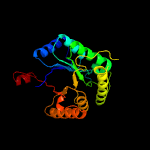
| 6 | c5w4zA_
|
|
 |
100.0 |
17 |
PDB header:flavoprotein
Chain: A: PDB Molecule:riboflavin lyase;
PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
|
|
|
|
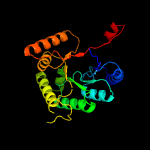
| 7 | d1lucb_
|
|
 |
100.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
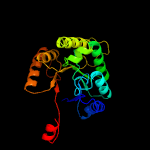
| 8 | d1luca_
|
|
 |
100.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
| 9 | c3raoB_
|
|
 |
100.0 |
21 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
|
|
|
| 10 | c1z69D_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:coenzyme f420-dependent n(5),n(10)-
PDBTitle: crystal structure of methylenetetrahydromethanopterin2 reductase (mer) in complex with coenzyme f420
|
|
|
|
| 11 | c6friD_
|
|
 |
100.0 |
13 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alkanal monooxygenase beta chain;
PDBTitle: structure of luxb from photobacterium leiognathi
|
|
|
|
| 12 | d1f07a_
|
|
 |
100.0 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 13 | d1ezwa_
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 14 | c5dqpA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:edta monooxygenase;
PDBTitle: edta monooxygenase (emoa) from chelativorans sp. bnc1
|
|
|
|
| 15 | c3b9nB_
|
|
 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkane monoxygenase;
PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
|
|
|
|
| 16 | c2i7gA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
|
|
|
|
| 17 | c2wgkA_
|
|
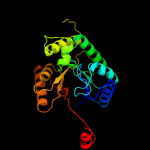 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,6-diketocamphane 1,6 monooxygenase;
PDBTitle: type ii baeyer-villiger monooxygenase oxygenating2 constituent of 3,6-diketocamphane 1,6 monooxygenase from3 pseudomonas putida
|
|
|
|
| 18 | c6ak1B_
|
|
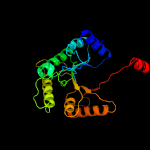 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dimethyl-sulfide monooxygenase;
PDBTitle: crystal structure of dmoa from hyphomicrobium sulfonivorans
|
|
|
|
| 19 | c3c8nB_
|
|
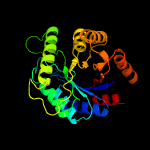 |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable f420-dependent glucose-6-phosphate dehydrogenase
PDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
|
|
|
|
| 20 | d1rhca_
|
|
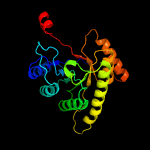 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 21 | d1nqka_ |
|
not modelled |
100.0 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
| 22 | c5wanA_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrimidine monooxygenase ruta;
PDBTitle: crystal structure of a flavoenzyme ruta in the pyrimidine catabolic2 pathway
|
|
|
| 23 | d1nfpa_ |
|
not modelled |
99.3 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 24 | d1fvpa_ |
|
not modelled |
96.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 25 | c3kwsB_ |
|
not modelled |
83.6 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:putative sugar isomerase;
PDBTitle: crystal structure of putative sugar isomerase (yp_001305149.1) from2 parabacteroides distasonis atcc 8503 at 1.68 a resolution
|
|
|
| 26 | c3ih1A_ |
|
not modelled |
73.9 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
|
|
| 27 | c3vniC_ |
|
not modelled |
68.0 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:xylose isomerase domain protein tim barrel;
PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
|
|
|
| 28 | c2hjpA_ |
|
not modelled |
61.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:phosphonopyruvate hydrolase;
PDBTitle: crystal structure of phosphonopyruvate hydrolase complex with2 phosphonopyruvate and mg++
|
|
|
| 29 | c3qy6A_ |
|
not modelled |
54.1 |
6 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase ywqe;
PDBTitle: crystal structures of ywqe from bacillus subtilis and cpsb from2 streptococcus pneumoniae, unique metal-dependent tyrosine3 phosphatases
|
|
|
| 30 | c5zfsA_ |
|
not modelled |
49.5 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:d-allulose-3-epimerase;
PDBTitle: crystal structure of arthrobacter globiformis m30 sugar epimerase2 which can produce d-allulose from d-fructose
|
|
|
| 31 | c3b8iF_ |
|
not modelled |
48.3 |
10 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|
| 32 | c6daoB_ |
|
not modelled |
48.2 |
11 |
PDB header:lyase
Chain: B: PDB Molecule:trans-o-hydroxybenzylidenepyruvate hydratase-aldolase;
PDBTitle: nahe wt selenomethionine
|
|
|
| 33 | d1oyaa_ |
|
not modelled |
47.3 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 34 | c2ekcA_ |
|
not modelled |
47.2 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: structural study of project id aq_1548 from aquifex aeolicus vf5
|
|
|
| 35 | c1ps9A_ |
|
not modelled |
45.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2,4-dienoyl-coa reductase;
PDBTitle: the crystal structure and reaction mechanism of e. coli 2,4-dienoyl2 coa reductase
|
|
|
| 36 | c4b5nA_ |
|
not modelled |
45.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: crystal structure of oxidized shewanella yellow enzyme 4 (sye4)
|
|
|
| 37 | c3eooL_ |
|
not modelled |
45.4 |
16 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
|
|
| 38 | c5dxxA_ |
|
not modelled |
45.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:artemisinic aldehyde delta(11(13)) reductase;
PDBTitle: crystal structure of dbr2
|
|
|
| 39 | c5tchG_ |
|
not modelled |
44.0 |
26 |
PDB header:lyase
Chain: G: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
| 40 | d1e0ta2 |
|
not modelled |
43.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Pyruvate kinase |
|
|
| 41 | c4tmcB_ |
|
not modelled |
41.6 |
25 |
PDB header:flavoprotein
Chain: B: PDB Molecule:old yellow enzyme;
PDBTitle: crystal structure of old yellow enzyme from candida macedoniensis2 aku4588 complexed with p-hydroxybenzaldehyde
|
|
|
| 42 | c4a3uB_ |
|
not modelled |
41.2 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadh\:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of the old yellow enzyme homologue from zymomonas mobilis2 (ncr)
|
|
|
| 43 | c1zlpA_ |
|
not modelled |
40.9 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
|
|
| 44 | d1gwja_ |
|
not modelled |
40.7 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 45 | c2gq8A_ |
|
not modelled |
40.3 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, fmn-binding;
PDBTitle: structure of sye1, an oye homologue from s. ondeidensis, in complex2 with p-hydroxyacetophenone
|
|
|
| 46 | c3dcpB_ |
|
not modelled |
38.5 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:histidinol-phosphatase;
PDBTitle: crystal structure of the putative histidinol phosphatase hisk from2 listeria monocytogenes. northeast structural genomics consortium3 target lmr141.
|
|
|
| 47 | d1q45a_ |
|
not modelled |
38.5 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 48 | c6daqA_ |
|
not modelled |
38.2 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:phdj;
PDBTitle: phdj bound to substrate intermediate
|
|
|
| 49 | c3l5aA_ |
|
not modelled |
38.1 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh/flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of a probable nadh-dependent flavin oxidoreductase2 from staphylococcus aureus
|
|
|
| 50 | d1vjia_ |
|
not modelled |
38.0 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 51 | d1vyra_ |
|
not modelled |
37.5 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 52 | c4jicB_ |
|
not modelled |
37.2 |
22 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gtn reductase;
PDBTitle: glycerol trinitrate reductase nera from agrobacterium radiobacter
|
|
|
| 53 | c1bplA_ |
|
not modelled |
37.2 |
11 |
PDB header:glycosyltransferase
Chain: A: PDB Molecule:alpha-1,4-glucan-4-glucanohydrolase;
PDBTitle: glycosyltransferase
|
|
|
| 54 | c5ocsB_ |
|
not modelled |
37.1 |
4 |
PDB header:flavoprotein
Chain: B: PDB Molecule:putative nadh-depentdent flavin oxidoreductase;
PDBTitle: ene-reductase (er/oye) from ralstonia (cupriavidus) metallidurans
|
|
|
| 55 | d1muma_ |
|
not modelled |
36.8 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 56 | c3gkaB_ |
|
not modelled |
36.3 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: crystal structure of n-ethylmaleimidine reductase from2 burkholderia pseudomallei
|
|
|
| 57 | c2ze3A_ |
|
not modelled |
36.0 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
|
|
| 58 | d1icpa_ |
|
not modelled |
34.9 |
25 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 59 | c2x7vA_ |
|
not modelled |
34.9 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
|
|
|
| 60 | c3e38A_ |
|
not modelled |
34.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:two-domain protein containing predicted php-like metal-
PDBTitle: crystal structure of a two-domain protein containing predicted php-2 like metal-dependent phosphoesterase (bvu_3505) from bacteroides3 vulgatus atcc 8482 at 2.20 a resolution
|
|
|
| 61 | c3fa4D_ |
|
not modelled |
34.3 |
13 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 62 | d1oy0a_ |
|
not modelled |
34.1 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Ketopantoate hydroxymethyltransferase PanB |
|
|
| 63 | c4rnxA_ |
|
not modelled |
34.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase 1;
PDBTitle: k154 circular permutation of old yellow enzyme
|
|
|
| 64 | d1djqa1 |
|
not modelled |
33.9 |
35 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 65 | c3ag5A_ |
|
not modelled |
33.0 |
23 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantothenate synthetase from staphylococcus2 aureus
|
|
|
| 66 | d1xp3a1 |
|
not modelled |
33.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:Endonuclease IV |
|
|
| 67 | d1z41a1 |
|
not modelled |
32.8 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 68 | c4df2A_ |
|
not modelled |
32.7 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph dehydrogenase;
PDBTitle: p. stipitis oye2.6 complexed with p-chlorophenol
|
|
|
| 69 | c2qiwA_ |
|
not modelled |
32.5 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
|
|
| 70 | c4qnwA_ |
|
not modelled |
31.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chanoclavine-i aldehyde reductase;
PDBTitle: crystal structure of easa, an old yellow enzyme from aspergillus2 fumigatus
|
|
|
| 71 | c3bc9A_ |
|
not modelled |
31.4 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:alpha amylase, catalytic region;
PDBTitle: alpha-amylase b in complex with acarbose
|
|
|
| 72 | c6mywA_ |
|
not modelled |
30.6 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: gluconobacter ene-reductase (gluer) mutant - t36a
|
|
|
| 73 | c2p0oA_ |
|
not modelled |
30.1 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein duf871;
PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
|
|
|
| 74 | c3vndD_ |
|
not modelled |
29.7 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha-subunit from the2 psychrophile shewanella frigidimarina k14-2
|
|
|
| 75 | c2vk2A_ |
|
not modelled |
29.3 |
14 |
PDB header:transport protein
Chain: A: PDB Molecule:abc transporter periplasmic-binding protein ytfq;
PDBTitle: crystal structure of a galactofuranose binding protein
|
|
|
| 76 | c3kruC_ |
|
not modelled |
29.1 |
25 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nadh:flavin oxidoreductase/nadh oxidase;
PDBTitle: crystal structure of the thermostable old yellow enzyme from2 thermoanaerobacter pseudethanolicus e39
|
|
|
| 77 | c4rnvD_ |
|
not modelled |
28.8 |
15 |
PDB header:oxidoreductase/inhibitor
Chain: D: PDB Molecule:nadph dehydrogenase 1;
PDBTitle: g303 circular permutation of old yellow enzyme with the inhibitor p-2 hydroxybenzaldehyde
|
|
|
| 78 | d1x7fa2 |
|
not modelled |
28.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Outer surface protein, N-terminal domain |
|
|
| 79 | c3dz1A_ |
|
not modelled |
28.3 |
10 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from2 rhodopseudomonas palustris at 1.87a resolution
|
|
|
| 80 | d1m5wa_ |
|
not modelled |
27.7 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Pyridoxine 5'-phosphate synthase
Family:Pyridoxine 5'-phosphate synthase |
|
|
| 81 | c3innB_ |
|
not modelled |
27.4 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of pantoate-beta-alanine-ligase in complex2 with atp at low occupancy at 2.1 a resolution
|
|
|
| 82 | d1s2wa_ |
|
not modelled |
27.3 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 83 | c2zvrA_ |
|
not modelled |
27.0 |
6 |
PDB header:isomerase
Chain: A: PDB Molecule:uncharacterized protein tm_0416;
PDBTitle: crystal structure of a d-tagatose 3-epimerase-related protein from2 thermotoga maritima
|
|
|
| 84 | c5epdA_ |
|
not modelled |
26.8 |
20 |
PDB header:oxidorecctase
Chain: A: PDB Molecule:glycerol trinitrate reductase;
PDBTitle: crystal structure of glycerol trinitrate reductase xdpb from2 agrobacterium sp. r89-1 (apo form)
|
|
|
| 85 | c2ejcA_ |
|
not modelled |
26.6 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:pantoate--beta-alanine ligase;
PDBTitle: crystal structure of pantoate--beta-alanine ligase (panc)2 from thermotoga maritima
|
|
|
| 86 | c2hk1D_ |
|
not modelled |
26.0 |
13 |
PDB header:isomerase
Chain: D: PDB Molecule:d-psicose 3-epimerase;
PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
|
|
|
| 87 | d1uoua2 |
|
not modelled |
25.1 |
17 |
Fold:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Superfamily:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain
Family:Nucleoside phosphorylase/phosphoribosyltransferase catalytic domain |
|
|
| 88 | c4ot7A_ |
|
not modelled |
25.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of a variant of ncr from zymomonas mobilis
|
|
|
| 89 | c3gk0H_ |
|
not modelled |
24.8 |
23 |
PDB header:transferase
Chain: H: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: crystal structure of pyridoxal phosphate biosynthetic protein from2 burkholderia pseudomallei
|
|
|
| 90 | c2yb1A_ |
|
not modelled |
24.6 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:amidohydrolase;
PDBTitle: structure of an amidohydrolase from chromobacterium violaceum (efi2 target efi-500202) with bound mn, amp and phosphate.
|
|
|
| 91 | d1ua7a2 |
|
not modelled |
22.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 92 | d1hl2a_ |
|
not modelled |
22.9 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 93 | d1goxa_ |
|
not modelled |
22.8 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 94 | c6k0aC_ |
|
not modelled |
22.7 |
0 |
PDB header:rna binding protein/rna
Chain: C: PDB Molecule:ribonuclease p protein component 3;
PDBTitle: cryo-em structure of an archaeal ribonuclease p
|
|
|
| 95 | c2a7nA_ |
|
not modelled |
22.6 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l(+)-mandelate dehydrogenase;
PDBTitle: crystal structure of the g81a mutant of the active chimera of (s)-2 mandelate dehydrogenase
|
|
|
| 96 | d1ud2a2 |
|
not modelled |
22.6 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 97 | d2gjpa2 |
|
not modelled |
22.6 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 98 | c3khjE_ |
|
not modelled |
22.1 |
10 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:inosine-5-monophosphate dehydrogenase;
PDBTitle: c. parvum inosine monophosphate dehydrogenase bound by inhibitor c64
|
|
|
| 99 | d1ihoa_ |
|
not modelled |
21.9 |
17 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC) |
|
|
| 100 | c5dlcC_ |
|
not modelled |
21.4 |
20 |
PDB header:transferase
Chain: C: PDB Molecule:pyridoxine 5'-phosphate synthase;
PDBTitle: x-ray crystal structure of a pyridoxine 5-prime-phosphate synthase2 from pseudomonas aeruginosa
|
|
|
| 101 | c5kwvA_ |
|
not modelled |
21.4 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:pantothenate synthetase;
PDBTitle: crystal structure of a pantoate-beta-alanine ligase from neisseria2 gonorrhoeae with bound amppnp
|
|
|
| 102 | c6de6B_ |
|
not modelled |
21.2 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:histamine dehydrogenase;
PDBTitle: 2.1 a resolution structure of histamine dehydrogenase from rhizobium2 sp. 4-9
|
|
|
| 103 | d1ujqa_ |
|
not modelled |
21.1 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 104 | c3obeB_ |
|
not modelled |
20.4 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:sugar phosphate isomerase/epimerase;
PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_3400)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
|
|
|
| 105 | c6agzA_ |
|
not modelled |
20.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:old yellow enzyme;
PDBTitle: crystal structure of old yellow enzyme from pichia sp. aku4542
|
|
|
























































































































































































