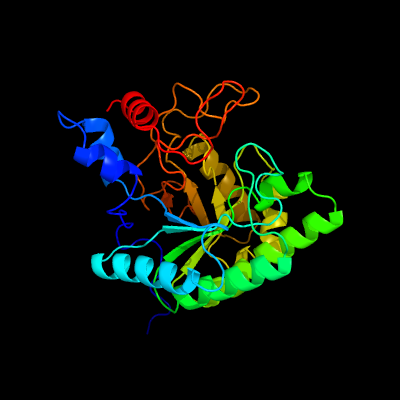
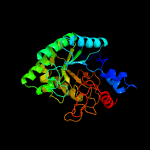
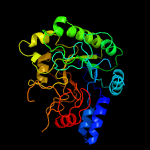
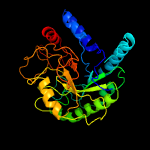
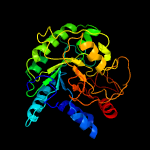
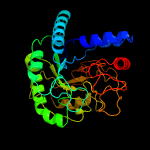
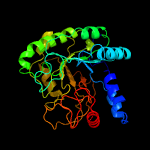
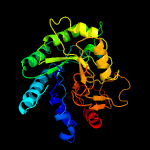
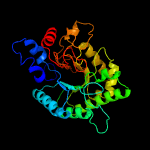
1 d1uoza_
100.0
99
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
2 c5xyhA_
100.0
34
PDB header: hydrolaseChain: A: PDB Molecule: cbsa;PDBTitle: crystal structure of catalytic domain of 1,4-beta-cellobiosidase2 (cbsa) from xanthomonas oryzae pv. oryzae
3 c3vohA_
100.0
33
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: cccel6a catalytic domain complexed with cellobiose
4 d1qjwa_
100.0
30
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
5 c5xczA_
100.0
30
PDB header: hydrolaseChain: A: PDB Molecule: glucanase;PDBTitle: structure of the cellobiohydrolase cel6a from phanerochaete2 chrysosporium in complex with cellobiose at 2.1 angstrom
6 d1oc7a_
100.0
30
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
7 d1dysa_
100.0
30
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
8 c3a64A_
100.0
29
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: crystal structure of cccel6c, a glycoside hydrolase family 62 enzyme, from coprinopsis cinerea
9 c4b4fB_
100.0
34
PDB header: hydrolaseChain: B: PDB Molecule: beta-1,4-exocellulase;PDBTitle: thermobifida fusca cel6b(e3) co-crystallized with cellobiose
10 c5jx6C_
100.0
31
PDB header: hydrolaseChain: C: PDB Molecule: glucanase;PDBTitle: gh6 orpinomyces sp. y102 enzyme
11 c6faoA_
100.0
48
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase family 6;PDBTitle: discovery and characterization of a thermostable gh6 endoglucanase2 from a compost metagenome
12 d2bodx1
100.0
43
Fold: 7-stranded beta/alpha barrelSuperfamily: Glycosyl hydrolases family 6, cellulasesFamily: Glycosyl hydrolases family 6, cellulases
13 c4w86B_
61.6
11
PDB header: hydrolaseChain: B: PDB Molecule: xyloglucan-specific endo-beta-1,4-glucanase;PDBTitle: crystal structure of xeg5a, a gh5 xyloglucan-specific endo-beta-1,4-2 glucanase from ruminal metagenomic library, in complex with glucose3 and tris
14 c4w7wA_
56.5
12
PDB header: hydrolaseChain: A: PDB Molecule: cellulase;PDBTitle: high-resolution structure of xaccel5a in complex with cellopentaose
15 c3w0kA_
50.9
14
PDB header: hydrolaseChain: A: PDB Molecule: bifunctional endomannanase/endoglucanase;PDBTitle: crystal structure of a glycoside hydrolase
16 c4lx4D_
50.1
10
PDB header: hydrolaseChain: D: PDB Molecule: endoglucanase(endo-1,4-beta-glucanase)protein;PDBTitle: crystal structure determination of pseudomonas stutzeri endoglucanase2 cel5a using a twinned data set
17 c5tnvA_
47.2
13
PDB header: isomeraseChain: A: PDB Molecule: ap endonuclease, family protein 2;PDBTitle: crystal structure of a xylose isomerase-like tim barrel protein from2 mycobacterium smegmatis in complex with magnesium
18 c3ne8A_
46.1
19
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase;PDBTitle: the crystal structure of a domain from n-acetylmuramoyl-l-alanine2 amidase of bartonella henselae str. houston-1
19 c4ee9A_
43.7
8
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: crystal structure of the rbcel1 endo-1,4-glucanase
20 c3u7bB_
40.4
20
PDB header: hydrolaseChain: B: PDB Molecule: endo-1,4-beta-xylanase;PDBTitle: a new crystal structure of a fusarium oxysporum gh10 xylanase reveals2 the presence of an extended loop on top of the catalytic cleft
21 d1d8wa_
not modelled
39.1
19
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: L-rhamnose isomerase
22 d2pw4a1
not modelled
37.2
50
Fold: Jann2411-likeSuperfamily: Jann2411-likeFamily: Jann2411-like
23 d1tz9a_
not modelled
36.7
10
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: UxuA-like
24 d3byqa1
not modelled
35.5
35
Fold: Bacillus chorismate mutase-likeSuperfamily: BB2672-likeFamily: BB2672-like
25 d2pb1a1
not modelled
35.2
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
26 c3obeB_
not modelled
33.8
7
PDB header: isomeraseChain: B: PDB Molecule: sugar phosphate isomerase/epimerase;PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_3400)2 from parabacteroides distasonis atcc 8503 at 1.70 a resolution
27 c3ndyA_
not modelled
33.4
10
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase d;PDBTitle: the structure of the catalytic and carbohydrate binding domain of2 endoglucanase d from clostridium cellulovorans
28 c4nf7A_
not modelled
33.3
9
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-glucanase cel5c;PDBTitle: crystal structure of the gh5 family catalytic domain of endo-1,4-beta-2 glucanase cel5c from butyrivibrio proteoclasticus.
29 c5w7dA_
not modelled
31.4
21
PDB header: hydrolaseChain: A: PDB Molecule: acyloxyacyl hydrolase;PDBTitle: murine acyloxyacyl hydrolase (aoah), s262a mutant
30 d1bxba_
not modelled
30.6
13
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
31 c4yztA_
not modelled
30.2
11
PDB header: hydrolaseChain: A: PDB Molecule: cellulose hydrolase;PDBTitle: crystal structure of a tri-modular gh5 (subfamily 4) endo-beta-1, 4-2 glucanase from bacillus licheniformis complexed with cellotetraose
32 c3vniC_
not modelled
29.9
10
PDB header: isomeraseChain: C: PDB Molecule: xylose isomerase domain protein tim barrel;PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
33 c5c5gA_
not modelled
29.8
15
PDB header: hydrolaseChain: A: PDB Molecule: spherulin-4;PDBTitle: crystal structure of aspergillus clavatus sph3
34 c2qtpA_
not modelled
29.6
36
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf1185 family protein (spo0826) from2 silicibacter pomeroyi dss-3 at 2.10 a resolution
35 d2qtpa1
not modelled
29.6
36
Fold: Bacillus chorismate mutase-likeSuperfamily: BB2672-likeFamily: BB2672-like
36 d1vjza_
not modelled
29.1
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
37 c4m6iA_
not modelled
28.5
33
PDB header: hydrolaseChain: A: PDB Molecule: peptidoglycan amidase rv3717;PDBTitle: structure of the reduced, zn-bound form of mycobacterium tuberculosis2 peptidoglycan amidase rv3717
38 c3anyB_
not modelled
27.4
28
PDB header: lyaseChain: B: PDB Molecule: ethanolamine ammonia-lyase light chain;PDBTitle: crystal structure of ethanolamine ammonia-lyase from escherichia coli2 complexed with cn-cbl and (r)-2-amino-1-propanol
39 c3absD_
not modelled
27.4
28
PDB header: lyaseChain: D: PDB Molecule: ethanolamine ammonia-lyase light chain;PDBTitle: crystal structure of ethanolamine ammonia-lyase from escherichia coli2 complexed with adeninylpentylcobalamin and ethanolamine
40 c5gnnA_
not modelled
27.3
25
PDB header: lipid binding proteinChain: A: PDB Molecule: lipid binding protein;PDBTitle: crystal structure of lipid binding protein nakanori at 1.6a
41 c3qxbB_
not modelled
24.9
10
PDB header: isomeraseChain: B: PDB Molecule: putative xylose isomerase;PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
42 c6gl2A_
not modelled
23.9
8
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase, family gh5;PDBTitle: structure of zgengagh5_4 wild type at 1.2 angstrom resolution
43 c4eisA_
not modelled
23.5
28
PDB header: oxidoreductaseChain: A: PDB Molecule: polysaccharide monooxygenase-3;PDBTitle: structural basis for substrate targeting and catalysis by fungal2 polysaccharide monooxygenases (pmo-3)
44 c3ktcB_
not modelled
23.0
15
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
45 c3p14C_
not modelled
22.5
13
PDB header: isomeraseChain: C: PDB Molecule: l-rhamnose isomerase;PDBTitle: crystal structure of l-rhamnose isomerase with a novel high thermo-2 stability from bacillus halodurans
46 c5hosA_
not modelled
22.2
10
PDB header: hydrolaseChain: A: PDB Molecule: cellulase;PDBTitle: crystal structure of the endo-beta-1,4-glucanase xac0029 from2 xanthomonas axonopodis pv. citri
47 c2zunB_
not modelled
21.1
12
PDB header: hydrolaseChain: B: PDB Molecule: 458aa long hypothetical endo-1,4-beta-glucanase;PDBTitle: functional analysis of hyperthermophilic endocellulase from2 the archaeon pyrococcus horikoshii
48 c5hmqE_
not modelled
20.5
21
PDB header: lyaseChain: E: PDB Molecule: 4-hydroxyphenylpyruvate dioxygenase;PDBTitle: xylose isomerase-like tim barrel/4-hydroxyphenylpyruvate dioxygenase2 fusion protein
49 c4d7uB_
not modelled
19.2
39
PDB header: oxidoreductaseChain: B: PDB Molecule: endoglucanase ii;PDBTitle: the structure of the catalytic domain of nclpmo9c from the filamentous2 fungus neurospora crassa
50 c4eirB_
not modelled
19.1
33
PDB header: oxidoreductaseChain: B: PDB Molecule: polysaccharide monooxygenase-2;PDBTitle: structural basis for substrate targeting and catalysis by fungal2 polysaccharide monooxygenases (pmo-2)
51 c4htyA_
not modelled
18.9
12
PDB header: hydrolaseChain: A: PDB Molecule: cellulase;PDBTitle: crystal structure of a metagenome-derived cellulase cel5a
52 c3qayC_
not modelled
17.9
23
PDB header: lyaseChain: C: PDB Molecule: endolysin;PDBTitle: catalytic domain of cd27l endolysin targeting clostridia difficile
53 d1hjsa_
not modelled
17.9
12
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
54 c2dfxI_
not modelled
17.4
18
PDB header: hydrolaseChain: I: PDB Molecule: colicin-e5 immunity protein;PDBTitle: crystal structure of the carboxy terminal domain of colicin2 e5 complexed with its inhibitor
55 c4ntqB_
not modelled
17.2
23
PDB header: toxinChain: B: PDB Molecule: ecl cdii;PDBTitle: cdia-ct/cdii toxin and immunity complex from enterobacter cloacae
56 d1ecea_
not modelled
17.0
12
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
57 c5a94B_
not modelled
16.8
15
PDB header: hydrolaseChain: B: PDB Molecule: putative retaining b-glycosidase;PDBTitle: crystal structure of beta-glucanase sdgluc5_26a from saccharophagus2 degradans in complex with tetrasaccharide a, form 1
58 c2hk1D_
not modelled
16.2
11
PDB header: isomeraseChain: D: PDB Molecule: d-psicose 3-epimerase;PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
59 d1xima_
not modelled
15.9
12
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
60 c4rl4B_
not modelled
15.7
19
PDB header: hydrolaseChain: B: PDB Molecule: gtp cyclohydrolase-2;PDBTitle: crystal structure of gtp cyclohydrolase ii from helicobacter pylori2 26695
61 c5emiA_
not modelled
15.6
13
PDB header: hydrolaseChain: A: PDB Molecule: cell wall hydrolase/autolysin;PDBTitle: n-acetylmuramoyl-l-alanine amidase amic2 of nostoc punctiforme
62 d2ifxa1
not modelled
15.4
55
Fold: Ferredoxin-likeSuperfamily: Dimeric alpha+beta barrelFamily: MmlI-like
63 c5dcjA_
not modelled
15.4
22
PDB header: transferaseChain: A: PDB Molecule: osmolarity sensor protein envz;PDBTitle: structure of the envz periplasmic domain with chaps reveals the2 mechanism of porin inactivation by bile salts
64 c3zmrA_
not modelled
15.3
11
PDB header: hydrolaseChain: A: PDB Molecule: cellulase (glycosyl hydrolase family 5);PDBTitle: bacteroides ovatus gh5 xyloglucanase in complex with a xxxg2 heptasaccharide
65 c5nltD_
not modelled
15.1
33
PDB header: oxidoreductaseChain: D: PDB Molecule: cvaa9a;PDBTitle: cvaa9a
66 c4b5qA_
not modelled
15.0
28
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase family 61 protein d;PDBTitle: the lytic polysaccharide monooxygenase gh61d structure from the2 basidiomycota fungus phanerochaete chrysosporium
67 c2rngA_
not modelled
14.9
40
PDB header: antimicrobial proteinChain: A: PDB Molecule: big defensin;PDBTitle: solution structure of big defensin
68 d1bqca_
not modelled
14.7
14
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
69 c3aysA_
not modelled
14.6
13
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: gh5 endoglucanase from a ruminal fungus in complex with cellotriose
70 c5aciA_
not modelled
14.5
39
PDB header: oxidoreductaseChain: A: PDB Molecule: lytic polysaccharide monooxygenase;PDBTitle: x-ray structure of lpmo
71 c4binA_
not modelled
14.3
31
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase amic;PDBTitle: crystal structure of the e. coli n-acetylmuramoyl-l-alanine amidase2 amic
72 d1edga_
not modelled
14.3
11
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
73 c2yetB_
not modelled
14.1
22
PDB header: hydrolaseChain: B: PDB Molecule: gh61 isozyme a;PDBTitle: thermoascus gh61 isozyme a
74 c5fohA_
not modelled
13.9
33
PDB header: oxidoreductaseChain: A: PDB Molecule: polysaccharide monooxygenase;PDBTitle: crystal structure of the catalytic domain of nclpmo9a
75 d1vpxa_
not modelled
13.8
17
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
76 d1ffvb2
not modelled
13.3
14
Fold: Molybdenum cofactor-binding domainSuperfamily: Molybdenum cofactor-binding domainFamily: Molybdenum cofactor-binding domain
77 c3ejaB_
not modelled
13.1
25
PDB header: unknown functionChain: B: PDB Molecule: protein gh61e;PDBTitle: magnesium-bound glycoside hydrolase 61 isoform e from thielavia2 terrestris
78 c2e12B_
not modelled
13.1
36
PDB header: translationChain: B: PDB Molecule: hypothetical protein xcc3642;PDBTitle: the crystal structure of xc5848 from xanthomonas campestris2 adopting a novel variant of sm-like motif
79 c4tufC_
not modelled
13.0
12
PDB header: hydrolaseChain: C: PDB Molecule: major extracellular endoglucanase;PDBTitle: catalytic domain of the major endoglucanase from xanthomonas2 campestris pv. campestris
80 d1sqwa1
not modelled
12.9
44
Fold: PUA domain-likeSuperfamily: PUA domain-likeFamily: PUA domain
81 c5o2xA_
not modelled
12.7
22
PDB header: oxidoreductaseChain: A: PDB Molecule: glycoside hydrolase family 61;PDBTitle: extended catalytic domain of h. jecorina lpmo9a a.k.a eg4
82 c4x0vH_
not modelled
12.5
11
PDB header: hydrolaseChain: H: PDB Molecule: beta-1,3-1,4-glucanase;PDBTitle: structure of a gh5 family lichenase from caldicellulosiruptor sp. f32
83 d2djka1
not modelled
12.2
10
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: PDI-like
84 c2j8fA_
not modelled
12.1
24
PDB header: hydrolaseChain: A: PDB Molecule: lysozyme;PDBTitle: crystal structure of the modular cpl-1 endolysin complexed with a2 peptidoglycan analogue (e94q mutant in complex with a disaccharide-3 pentapeptide)
85 c3bj5A_
not modelled
12.0
10
PDB header: isomeraseChain: A: PDB Molecule: protein disulfide-isomerase;PDBTitle: alternative conformations of the x region of human protein disulphide-2 isomerase modulate exposure of the substrate binding b' domain
86 d1xova2
not modelled
11.5
30
Fold: Phosphorylase/hydrolase-likeSuperfamily: Zn-dependent exopeptidasesFamily: N-acetylmuramoyl-L-alanine amidase-like
87 c5fipA_
not modelled
11.4
9
PDB header: hydrolaseChain: A: PDB Molecule: gh5 cellulase;PDBTitle: discovery and characterization of a novel thermostable and2 highly halotolerant gh5 cellulase from an icelandic hot3 spring isolate
88 c3hynA_
not modelled
11.2
16
PDB header: signaling proteinChain: A: PDB Molecule: putative signal transduction protein;PDBTitle: crystal structure of a putative signal transduction protein2 (eubrec_0645) from eubacterium rectale atcc 33656 at 1.20 a3 resolution
89 c4k3zA_
not modelled
10.9
10
PDB header: oxidoreductaseChain: A: PDB Molecule: d-erythrulose 4-phosphate dehydrogenase;PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
90 d1bxca_
not modelled
10.9
15
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
91 d1x3ha2
not modelled
10.8
10
Fold: Glucocorticoid receptor-like (DNA-binding domain)Superfamily: Glucocorticoid receptor-like (DNA-binding domain)Family: LIM domain
92 c3wugA_
not modelled
10.8
12
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-xylanase a;PDBTitle: the mutant crystal structure of b-1,4-xylanase (xynas9_v43p/g44e) with2 xylobiose from streptomyces sp. 9
93 c4rn7A_
not modelled
10.8
40
PDB header: hydrolaseChain: A: PDB Molecule: n-acetylmuramoyl-l-alanine amidase;PDBTitle: the crystal structure of n-acetylmuramoyl-l-alanine amidase from2 clostridium difficile 630
94 c2qw5B_
not modelled
10.6
11
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase-like tim barrel;PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
95 c5dlcC_
not modelled
10.4
18
PDB header: transferaseChain: C: PDB Molecule: pyridoxine 5'-phosphate synthase;PDBTitle: x-ray crystal structure of a pyridoxine 5-prime-phosphate synthase2 from pseudomonas aeruginosa
96 d1s99a_
not modelled
10.3
33
Fold: Ferredoxin-likeSuperfamily: MTH1187/YkoF-likeFamily: Putative thiamin/HMP-binding protein YkoF
97 c3qr3B_
not modelled
10.0
11
PDB header: hydrolaseChain: B: PDB Molecule: endoglucanase eg-ii;PDBTitle: crystal structure of cel5a (eg2) from hypocrea jecorina (trichoderma2 reesei)
98 c3excX_
not modelled
10.0
22
PDB header: hydrolaseChain: X: PDB Molecule: uncharacterized protein;PDBTitle: structure of the rna'se sso8090 from sulfolobus solfataricus
99 c2inrA_
not modelled
9.8
11
PDB header: isomeraseChain: A: PDB Molecule: dna topoisomerase 4 subunit a;PDBTitle: crystal structure of a 59 kda fragment of topoisomerase iv subunit a2 (grla) from staphylococcus aureus