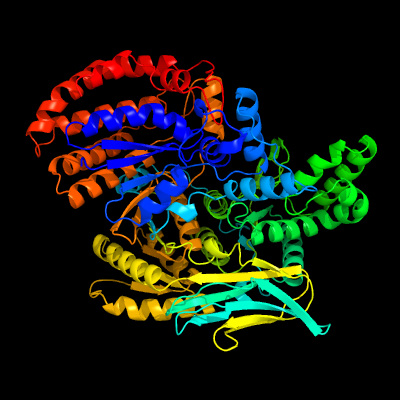
| 1 | d1itwa_
|
|
 |
100.0 |
61 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Monomeric isocitrate dehydrogenase |
|
|
|
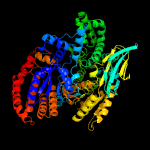
| 2 | c6g3uA_
|
|
 |
100.0 |
68 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: structure of pseudomonas aeruginosa isocitrate dehydrogenase, idh
|
|
|
|
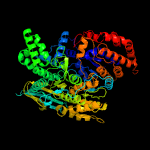
| 3 | c2b0tA_
|
|
 |
100.0 |
58 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadp isocitrate dehydrogenase;
PDBTitle: structure of monomeric nadp isocitrate dehydrogenase
|
|
|
|
| 4 | d1lwda_
|
|
 |
99.3 |
15 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 5 | c1zorB_
|
|
 |
99.1 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile thermotoga maritima
|
|
|
|
| 6 | c3us8A_
|
|
 |
98.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of an isocitrate dehydrogenase from sinorhizobium2 meliloti 1021
|
|
|
|
| 7 | c3blxL_
|
|
 |
97.8 |
17 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:isocitrate dehydrogenase [nad] subunit 2;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
|
|
|
| 8 | d1wpwa_
|
|
 |
97.8 |
16 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 9 | c3blxM_
|
|
 |
97.7 |
12 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:isocitrate dehydrogenase [nad] subunit 1;
PDBTitle: yeast isocitrate dehydrogenase (apo form)
|
|
|
|
| 10 | d1t0la_
|
|
 |
97.6 |
25 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 11 | c5grhB_
|
|
 |
97.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase [nad] subunit gamma,
PDBTitle: crystal structure of the alpha gamma heterodimer of human idh3 in2 complex with mg(2+)
|
|
|
|
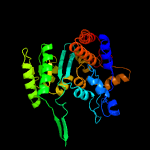
| 12 | c5hn6A_
|
|
 |
97.4 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of beta-decarboxylating dehydrogenase (tk0280) from2 thermococcus kodakarensis complexed with mn and 3-isopropylmalate
|
|
|
|
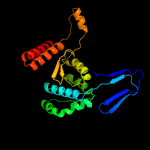
| 13 | c2uxqB_
|
|
 |
97.4 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase native;
PDBTitle: isocitrate dehydrogenase from the psychrophilic bacterium2 desulfotalea psychrophila: biochemical properties and3 crystal structure analysis
|
|
|
|
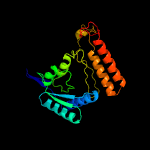
| 14 | c5grhA_
|
|
 |
97.3 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase [nad] subunit alpha,
PDBTitle: crystal structure of the alpha gamma heterodimer of human idh3 in2 complex with mg(2+)
|
|
|
|
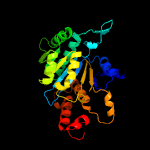
| 15 | c2e0cA_
|
|
 |
97.3 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:409aa long hypothetical nadp-dependent isocitrate
PDBTitle: crystal structure of isocitrate dehydrogenase from sulfolobus tokodaii2 strain7 at 2.0 a resolution
|
|
|
|
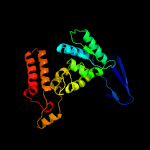
| 16 | c2qfyE_
|
|
 |
97.0 |
14 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: crystal structure of saccharomyces cerevesiae mitochondrial nadp(+)-2 dependent isocitrate dehydrogenase in complex with a-ketoglutarate
|
|
|
|
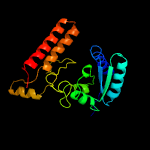
| 17 | c3fmxX_
|
|
 |
96.9 |
15 |
PDB header:oxidoreductase
Chain: X: PDB Molecule:tartrate dehydrogenase/decarboxylase;
PDBTitle: crystal structure of tartrate dehydrogenase from pseudomonas putida2 complexed with nadh
|
|
|
|
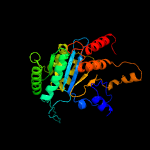
| 18 | d1g2ua_
|
|
 |
96.7 |
20 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
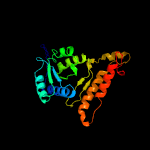
| 19 | d1w0da_
|
|
 |
96.6 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
|
| 20 | c2d1cB_
|
|
 |
96.6 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of tt0538 protein from thermus thermophilus hb8
|
|
|
|
| 21 | d1xaca_ |
|
not modelled |
96.1 |
23 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 22 | d1pb1a_ |
|
not modelled |
96.0 |
13 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 23 | c1x0lB_ |
|
not modelled |
95.7 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:homoisocitrate dehydrogenase;
PDBTitle: crystal structure of tetrameric homoisocitrate dehydrogenase from an2 extreme thermophile, thermus thermophilus
|
|
|
| 24 | c3ty3A_ |
|
not modelled |
95.6 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable homoisocitrate dehydrogenase;
PDBTitle: crystal structure of homoisocitrate dehydrogenase from2 schizosaccharomyces pombe bound to glycyl-glycyl-glycine
|
|
|
| 25 | c4aoyD_ |
|
not modelled |
95.4 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase [nadp];
PDBTitle: open ctidh. the complex structures of isocitrate dehydrogenase from2 clostridium thermocellum and desulfotalea psychrophila, support a new3 active site locking mechanism
|
|
|
| 26 | c2d4vD_ |
|
not modelled |
95.3 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: crystal structure of nad dependent isocitrate dehydrogenase2 from acidithiobacillus thiooxidans
|
|
|
| 27 | d1v53a1 |
|
not modelled |
94.9 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 28 | c3vl3A_ |
|
not modelled |
94.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: 3-isopropylmalate dehydrogenase from shewanella oneidensis mr-1 at 3402 mpa
|
|
|
| 29 | c1tyoA_ |
|
not modelled |
94.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: isocitrate dehydrogenase from the hyperthermophile aeropyrum pernix in2 complex with etheno-nadp
|
|
|
| 30 | d1hqsa_ |
|
not modelled |
94.4 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 31 | d1a05a_ |
|
not modelled |
94.1 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 32 | c4iwhA_ |
|
not modelled |
94.1 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of a 3-isopropylmalate dehydrogenase from2 burkholderia pseudomallei
|
|
|
| 33 | c2iv0A_ |
|
not modelled |
92.4 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:isocitrate dehydrogenase;
PDBTitle: thermal stability of isocitrate dehydrogenase from2 archaeoglobus fulgidus studied by crystal structure3 analysis and engineering of chimers
|
|
|
| 34 | d1vlca_ |
|
not modelled |
91.2 |
18 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 35 | d1cnza_ |
|
not modelled |
87.1 |
17 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 36 | d1cm7a_ |
|
not modelled |
85.1 |
19 |
Fold:Isocitrate/Isopropylmalate dehydrogenase-like
Superfamily:Isocitrate/Isopropylmalate dehydrogenase-like
Family:Dimeric isocitrate & isopropylmalate dehydrogenases |
|
|
| 37 | c3u1hA_ |
|
not modelled |
84.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of ipmdh from the last common ancestor of bacillus
|
|
|
| 38 | c3uduG_ |
|
not modelled |
83.7 |
16 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:3-isopropylmalate dehydrogenase;
PDBTitle: crystal structure of putative 3-isopropylmalate dehydrogenase from2 campylobacter jejuni
|
|
|
| 39 | d3ovwa_ |
|
not modelled |
83.1 |
25 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Glycosyl hydrolase family 7 catalytic core |
|
|
| 40 | c3r8wC_ |
|
not modelled |
81.4 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-isopropylmalate dehydrogenase 2, chloroplastic;
PDBTitle: structure of 3-isopropylmalate dehydrogenase isoform 2 from2 arabidopsis thaliana at 2.2 angstrom resolution
|
|
|
| 41 | d1ojja_ |
|
not modelled |
79.5 |
18 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Glycosyl hydrolase family 7 catalytic core |
|
|
| 42 | d1cooa_ |
|
not modelled |
74.2 |
20 |
Fold:SAM domain-like
Superfamily:C-terminal domain of RNA polymerase alpha subunit
Family:C-terminal domain of RNA polymerase alpha subunit |
|
|
| 43 | c3w7aD_ |
|
not modelled |
70.2 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:fmn-dependent nadh-azoreductase;
PDBTitle: crystal structure of azoreductase azrc fin complex with sulfone-2 modified azo dye acid red 88
|
|
|
| 44 | c4c90B_ |
|
not modelled |
65.2 |
23 |
PDB header:hydrolase
Chain: B: PDB Molecule:alpha-glucuronidase gh115;
PDBTitle: evidence that gh115 alpha-glucuronidase activity is2 dependent on conformational flexibility
|
|
|
| 45 | c3iydA_ |
|
not modelled |
55.9 |
20 |
PDB header:transcription/dna
Chain: A: PDB Molecule:dna-directed rna polymerase subunit alpha;
PDBTitle: three-dimensional em structure of an intact activator-dependent2 transcription initiation complex
|
|
|
| 46 | c3igsB_ |
|
not modelled |
49.7 |
15 |
PDB header:isomerase
Chain: B: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase 2;
PDBTitle: structure of the salmonella enterica n-acetylmannosamine-6-phosphate2 2-epimerase
|
|
|
| 47 | d1ee6a_ |
|
not modelled |
49.2 |
30 |
Fold:Single-stranded right-handed beta-helix
Superfamily:Pectin lyase-like
Family:Pectate lyase-like |
|
|
| 48 | c4u49B_ |
|
not modelled |
48.0 |
27 |
PDB header:lyase
Chain: B: PDB Molecule:pectate lyase;
PDBTitle: crystal structure of pectate lyase pel3 from pectobacterium2 carotovorum with two monomers in the a.u
|
|
|
| 49 | c4xnnA_ |
|
not modelled |
46.8 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:cellobiohydrolase chbi;
PDBTitle: crystal structure of a gh7 family cellobiohydrolase from daphnia pulex
|
|
|
| 50 | c3b90A_ |
|
not modelled |
44.6 |
29 |
PDB header:lyase
Chain: A: PDB Molecule:endo-pectate lyase;
PDBTitle: crystal structure of the catalytic domain of pectate lyase peli from2 erwinia chrysanthemi
|
|
|
| 51 | d1sxjd1 |
|
not modelled |
44.1 |
9 |
Fold:post-AAA+ oligomerization domain-like
Superfamily:post-AAA+ oligomerization domain-like
Family:DNA polymerase III clamp loader subunits, C-terminal domain |
|
|
| 52 | c5h6iC_ |
|
not modelled |
43.2 |
26 |
PDB header:toxin
Chain: C: PDB Molecule:protein b;
PDBTitle: crystal structure of gbs camp factor
|
|
|
| 53 | c3t9gB_ |
|
not modelled |
42.5 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:pectate lyase;
PDBTitle: the crystal structure of family 3 pectate lyase from2 caldicellulosiruptor bescii
|
|
|
| 54 | c3r07C_ |
|
not modelled |
42.1 |
7 |
PDB header:transferase
Chain: C: PDB Molecule:putative lipoate-protein ligase a subunit 2;
PDBTitle: structural analysis of an archaeal lipoylation system. a bi-partite2 lipoate protein ligase and its e2 lipoyl domain from thermoplasma3 acidophilum
|
|
|
| 55 | c3b4nB_ |
|
not modelled |
42.1 |
27 |
PDB header:lyase
Chain: B: PDB Molecule:endo-pectate lyase;
PDBTitle: crystal structure analysis of pectate lyase peli from2 erwinia chrysanthemi
|
|
|
| 56 | d1doqa_ |
|
not modelled |
41.2 |
21 |
Fold:SAM domain-like
Superfamily:C-terminal domain of RNA polymerase alpha subunit
Family:C-terminal domain of RNA polymerase alpha subunit |
|
|
| 57 | c4nz3A_ |
|
not modelled |
40.4 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:deacetylase da1;
PDBTitle: structure of vibrio cholerae chitin de-n-acetylase in complex with2 di(n-acetyl-d-glucosamine) (cbs) in p 21 21 21
|
|
|
| 58 | c4csiB_ |
|
not modelled |
38.8 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:cellulase;
PDBTitle: crystal structure of the thermostable cellobiohydrolase2 cel7a from the fungus humicola grisea var. thermoidea.
|
|
|
| 59 | d1lb2b_ |
|
not modelled |
37.9 |
19 |
Fold:SAM domain-like
Superfamily:C-terminal domain of RNA polymerase alpha subunit
Family:C-terminal domain of RNA polymerase alpha subunit |
|
|
| 60 | c4zmhA_ |
|
not modelled |
36.7 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a five-domain gh115 alpha-glucuronidase from the2 marine bacterium saccharophagus degradans 2-40t
|
|
|
| 61 | c1npyA_ |
|
not modelled |
35.2 |
17 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical shikimate 5-dehydrogenase-like protein hi0607;
PDBTitle: structure of shikimate 5-dehydrogenase-like protein hi0607
|
|
|
| 62 | c3tsjA_ |
|
not modelled |
34.3 |
14 |
PDB header:allergen, oxidoreductase
Chain: A: PDB Molecule:pollen allergen phl p 4;
PDBTitle: crystal structure of phl p 4, a grass pollen allergen with glucose2 dehydrogenase activity
|
|
|
| 63 | c5y2gA_ |
|
not modelled |
33.7 |
22 |
PDB header:toxin
Chain: A: PDB Molecule:maltose-binding periplasmic protein,protein b;
PDBTitle: structure of mbp tagged gbs camp
|
|
|
| 64 | c4toqC_ |
|
not modelled |
32.7 |
25 |
PDB header:hydrolase
Chain: C: PDB Molecule:class iii chitinase;
PDBTitle: crystal structure of class iii chitinase from pomegranate provides the2 insight into its metal storage capacity
|
|
|
| 65 | c6cg8A_ |
|
not modelled |
32.2 |
17 |
PDB header:dna binding protein/dna
Chain: A: PDB Molecule:upf0335 protein b7z12_12435;
PDBTitle: structure of c. crescentus gapr-dna
|
|
|
| 66 | c5kc1H_ |
|
not modelled |
31.8 |
60 |
PDB header:endocytosis
Chain: H: PDB Molecule:autophagy-related protein 38;
PDBTitle: structure of the c-terminal dimerization domain of atg38
|
|
|
| 67 | c1dvpA_ |
|
not modelled |
30.9 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine
PDBTitle: crystal structure of the vhs and fyve tandem domains of hrs,2 a protein involved in membrane trafficking and signal3 transduction
|
|
|
| 68 | c2gsjA_ |
|
not modelled |
30.7 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein ppl-2;
PDBTitle: cdna cloning and 1.75a crystal structure determination of ppl2, a2 novel chimerolectin from parkia platycephala seeds exhibiting3 endochitinolytic activity
|
|
|
| 69 | c6dxpD_ |
|
not modelled |
30.3 |
16 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:fmn-dependent nadh-azoreductase;
PDBTitle: the crystal structure of an fmn-dependent nadh-azoreductase from2 klebsiella pneumoniae
|
|
|
| 70 | c3q58A_ |
|
not modelled |
30.1 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:n-acetylmannosamine-6-phosphate 2-epimerase;
PDBTitle: structure of n-acetylmannosamine-6-phosphate epimerase from salmonella2 enterica
|
|
|
| 71 | c4hapB_ |
|
not modelled |
29.6 |
14 |
PDB header:hydrolase
Chain: B: PDB Molecule:gh7 family protein;
PDBTitle: crystal structure of a gh7 family cellobiohydrolase from limnoria2 quadripunctata in complex with cellobiose
|
|
|
| 72 | d1ebfa2 |
|
not modelled |
29.3 |
29 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:Homoserine dehydrogenase-like |
|
|
| 73 | d2v3ia1 |
|
not modelled |
29.0 |
15 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Glycosyl hydrolase family 7 catalytic core |
|
|
| 74 | d2obba1 |
|
not modelled |
28.8 |
29 |
Fold:HAD-like
Superfamily:HAD-like
Family:BT0820-like |
|
|
| 75 | c2maxA_ |
|
not modelled |
28.7 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:dna-directed rna polymerase subunit alpha;
PDBTitle: nmr structure of the rna polymerase alpha subunit c-terminal domain2 from helicobacter pylori
|
|
|
| 76 | c2q62A_ |
|
not modelled |
28.5 |
18 |
PDB header:flavoprotein
Chain: A: PDB Molecule:arsh;
PDBTitle: crystal structure of arsh from sinorhizobium meliloti
|
|
|
| 77 | c5f5tB_ |
|
not modelled |
28.4 |
28 |
PDB header:splicing
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the prp38-mfap1 complex of chaetomium2 thermophilum
|
|
|
| 78 | c2yg1A_ |
|
not modelled |
27.7 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:cellulose 1,4-beta-cellobiosidase;
PDBTitle: apo structure of cellobiohydrolase 1 (cel7a) from heterobasidion2 annosum
|
|
|
| 79 | c3bvkC_ |
|
not modelled |
27.7 |
14 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:ferritin;
PDBTitle: structural basis for the iron uptake mechanism of helicobacter pylori2 ferritin
|
|
|
| 80 | c4a54A_ |
|
not modelled |
27.2 |
19 |
PDB header:rna binding protein/hydrolase
Chain: A: PDB Molecule:edc3;
PDBTitle: structural basis of the dcp1:dcp2 mrna decapping complex activation2 by edc3 and scd6
|
|
|
| 81 | c4davA_ |
|
not modelled |
26.9 |
25 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:sugar fermentation stimulation protein homolog;
PDBTitle: the structure of pyrococcus furiosus sfsa in complex with dna
|
|
|
| 82 | d1uika2 |
|
not modelled |
26.9 |
9 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
|
|
| 83 | c1yunB_ |
|
not modelled |
26.4 |
29 |
PDB header:transferase
Chain: B: PDB Molecule:probable nicotinate-nucleotide
PDBTitle: crystal structure of nicotinic acid mononucleotide2 adenylyltransferase from pseudomonas aeruginosa
|
|
|
| 84 | d1fwxa2 |
|
not modelled |
26.2 |
15 |
Fold:7-bladed beta-propeller
Superfamily:Nitrous oxide reductase, N-terminal domain
Family:Nitrous oxide reductase, N-terminal domain |
|
|
| 85 | c4rpiA_ |
|
not modelled |
25.8 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:nicotinate-nucleotide adenylyltransferase;
PDBTitle: crystal structure of nicotinate mononucleotide adenylyltransferase2 from mycobacterium tuberculosis
|
|
|
| 86 | c2hpvA_ |
|
not modelled |
24.7 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fmn-dependent nadh-azoreductase;
PDBTitle: crystal structure of fmn-dependent azoreductase from enterococcus2 faecalis
|
|
|
| 87 | d2olra2 |
|
not modelled |
24.3 |
12 |
Fold:PEP carboxykinase N-terminal domain
Superfamily:PEP carboxykinase N-terminal domain
Family:PEP carboxykinase N-terminal domain |
|
|
| 88 | c2y9zB_ |
|
not modelled |
24.2 |
43 |
PDB header:transcription
Chain: B: PDB Molecule:iswi one complex protein 3;
PDBTitle: chromatin remodeling factor isw1a(del_atpase) in dna complex
|
|
|
| 89 | c3zyqA_ |
|
not modelled |
23.9 |
7 |
PDB header:signaling
Chain: A: PDB Molecule:hepatocyte growth factor-regulated tyrosine kinase
PDBTitle: crystal structure of the tandem vhs and fyve domains of hepatocyte2 growth factor-regulated tyrosine kinase substrate (hgs-hrs) at 1.48 a3 resolution
|
|
|
| 90 | c2fzvC_ |
|
not modelled |
23.9 |
14 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:putative arsenical resistance protein;
PDBTitle: crystal structure of an apo form of a flavin-binding protein from2 shigella flexneri
|
|
|
| 91 | d1fxza2 |
|
not modelled |
23.5 |
11 |
Fold:Double-stranded beta-helix
Superfamily:RmlC-like cupins
Family:Germin/Seed storage 7S protein |
|
|
| 92 | c3w8wA_ |
|
not modelled |
23.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative fad-dependent oxygenase encm;
PDBTitle: the crystal structure of encm
|
|
|
| 93 | c2xucA_ |
|
not modelled |
22.7 |
25 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitinase;
PDBTitle: natural product-guided discovery of a fungal chitinase inhibitor
|
|
|
| 94 | d1qopa_ |
|
not modelled |
22.4 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 95 | c3p0rA_ |
|
not modelled |
22.4 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:azoreductase;
PDBTitle: crystal structure of azoreductase from bacillus anthracis str. sterne
|
|
|
| 96 | c5zbaA_ |
|
not modelled |
22.0 |
45 |
PDB header:transferase/structural protein
Chain: A: PDB Molecule:dna damage response protein rtt109, putative;
PDBTitle: crystal structure of rtt109-asf1-h3-h4-coa complex
|
|
|
| 97 | c3c8mA_ |
|
not modelled |
22.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:homoserine dehydrogenase;
PDBTitle: crystal structure of homoserine dehydrogenase from thermoplasma2 volcanium
|
|
|
| 98 | d1juqa_ |
|
not modelled |
21.8 |
21 |
Fold:alpha-alpha superhelix
Superfamily:ENTH/VHS domain
Family:VHS domain |
|
|
| 99 | c4b16A_ |
|
not modelled |
21.4 |
31 |
PDB header:hydrolase
Chain: A: PDB Molecule:chitinase like lectin;
PDBTitle: crystal structure of tamarind chitinase like lectin (tcll) complexed2 with n-acetyl glucosamine (glcnac)
|
|
|
| 100 | d1zs6a1 |
|
not modelled |
21.1 |
17 |
Fold:Ferredoxin-like
Superfamily:Nucleoside diphosphate kinase, NDK
Family:Nucleoside diphosphate kinase, NDK |
|
|
| 101 | c3fseB_ |
|
not modelled |
21.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:two-domain protein containing dj-1/thij/pfpi-like and
PDBTitle: crystal structure of a two-domain protein containing dj-1/thij/pfpi-2 like and ferritin-like domains (ava_4496) from anabaena variabilis3 atcc 29413 at 1.90 a resolution
|
|
|
| 102 | c1vw48_ |
|
not modelled |
21.0 |
64 |
PDB header:ribosome
Chain: 8: PDB Molecule:54s ribosomal protein l13, mitochondrial;
PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
|
|
|
| 103 | d1krqa_ |
|
not modelled |
20.5 |
16 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ferritin |
|
|
| 104 | d1gpia_ |
|
not modelled |
20.4 |
15 |
Fold:Concanavalin A-like lectins/glucanases
Superfamily:Concanavalin A-like lectins/glucanases
Family:Glycosyl hydrolase family 7 catalytic core |
|
|
| 105 | c6cfxD_ |
|
not modelled |
20.4 |
21 |
PDB header:dna binding protein
Chain: D: PDB Molecule:upf0335 protein ase63_04290;
PDBTitle: bosea sp gapr solved in the presence of dna
|
|
|
| 106 | c3daoB_ |
|
not modelled |
20.1 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative phosphate;
PDBTitle: crystal structure of a putative phosphate (eubrec_1417) from2 eubacterium rectale at 1.80 a resolution
|
|
|