1 c4rqoB_
100.0
50
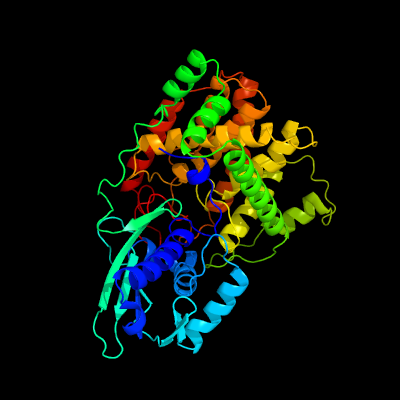
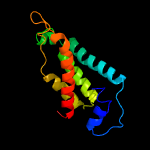
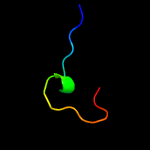
PDB header: lyaseChain: B: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure of l-serine dehydratase from legionella pneumophila
2 d2iafa1
100.0
42
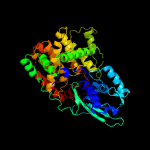
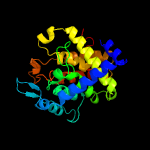
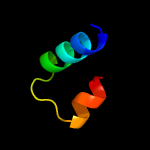
Fold: FwdE/GAPDH domain-likeSuperfamily: Serine metabolism enzymes domainFamily: Serine dehydratase beta chain-like
3 c1ygyA_
99.7
18
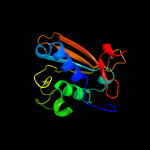
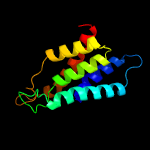
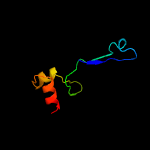
PDB header: oxidoreductaseChain: A: PDB Molecule: d-3-phosphoglycerate dehydrogenase;PDBTitle: crystal structure of d-3-phosphoglycerate dehydrogenase from2 mycobacterium tuberculosis
4 d1ygya4
99.4
20
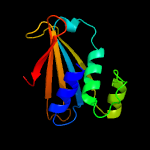
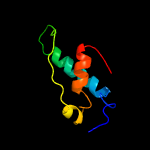
Fold: FwdE/GAPDH domain-likeSuperfamily: Serine metabolism enzymes domainFamily: SerA intervening domain-like
5 d1szqa_
97.6
20
Fold: 2-methylcitrate dehydratase PrpDSuperfamily: 2-methylcitrate dehydratase PrpDFamily: 2-methylcitrate dehydratase PrpD
6 c2hp0A_
97.1
15
PDB header: isomeraseChain: A: PDB Molecule: ids-epimerase;PDBTitle: crystal structure of iminodisuccinate epimerase
7 c5muxB_
96.5
17
PDB header: lyaseChain: B: PDB Molecule: 2-methylcitrate dehydratase;PDBTitle: crystal structure of 2-methylcitrate dehydratase (mmge) from bacillus2 subtilis.
8 c4zhjA_
48.9
22
PDB header: metal binding proteinChain: A: PDB Molecule: mg-chelatase subunit chlh;PDBTitle: crystal structure of the catalytic subunit of magnesium chelatase
9 c2m2rA_
46.6
75
PDB header: unknown functionChain: A: PDB Molecule: inhibitor cystine knot peptide mch-2;PDBTitle: solution structure of mch-2: a novel inhibitor cystine knot peptide2 from momordica charantia
10 c2w4eA_
45.6
14
PDB header: hydrolaseChain: A: PDB Molecule: mutt/nudix family protein;PDBTitle: structure of an n-terminally truncated nudix hydrolase2 dr2204 from deinococcus radiodurans
11 c3u4gA_
36.9
33
PDB header: transferaseChain: A: PDB Molecule: namn:dmb phosphoribosyltransferase;PDBTitle: the structure of cobt from pyrococcus horikoshii
12 c6b2zd_
35.5
24
PDB header: membrane proteinChain: D: PDB Molecule: atp synthase subunit c, mitochondrial;PDBTitle: cryo-em structure of the dimeric fo region of yeast mitochondrial atp2 synthase
13 c3j9eD_
35.4
21
PDB header: viral proteinChain: D: PDB Molecule: vp5;PDBTitle: atomic structure of a non-enveloped virus reveals ph sensors for a2 coordinated process of cell entry
14 c3s1sA_
30.5
35
PDB header: hydrolase, transferaseChain: A: PDB Molecule: restriction endonuclease bpusi;PDBTitle: characterization and crystal structure of the type iig restriction2 endonuclease bpusi
15 d1ug2a_
30.0
13
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: Myb/SANT domain
16 c5oynB_
27.4
22
PDB header: lyaseChain: B: PDB Molecule: dehydratase, ilvd/edd family;PDBTitle: crystal structure of d-xylonate dehydratase in holo-form
17 c5cagA_
26.1
27
PDB header: cell adhesionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a putative adhesin (bacova_02677) from2 bacteroides ovatus atcc 8483 at 3.00 a resolution (psi community3 target, nakayama)
18 d1no5a_
23.1
28
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase
19 d1rm6c1
22.8
12
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
20 d2i7pa1
22.5
25
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like
21 d1ffva1
not modelled
20.4
12
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
22 c2zfdB_
not modelled
20.1
25
PDB header: signaling protein/transferaseChain: B: PDB Molecule: putative uncharacterized protein t20l15_90;PDBTitle: the crystal structure of plant specific calcium binding protein atcbl22 in complex with the regulatory domain of atcipk14
23 c2i7pA_
not modelled
20.0
25
PDB header: transferaseChain: A: PDB Molecule: pantothenate kinase 3;PDBTitle: crystal structure of human pank3 in complex with accoa
24 d2i7na2
not modelled
19.1
21
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Fumble-like
25 c1yy3A_
not modelled
18.7
57
PDB header: isomeraseChain: A: PDB Molecule: s-adenosylmethionine:trna ribosyltransferase-PDBTitle: structure of s-adenosylmethionine:trna ribosyltransferase-2 isomerase (quea)
26 d1fh9a_
not modelled
18.1
8
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
27 c5xzoA_
not modelled
18.0
20
PDB header: hydrolaseChain: A: PDB Molecule: beta-xylanase;PDBTitle: crystal structure of gh10 xylanase xyl10c from bispora. sp mey-1
28 d1zxia1
not modelled
17.8
4
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
29 d1v97a1
not modelled
17.5
4
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
30 d1ncfb3
not modelled
16.2
40
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
31 c5mrjA_
not modelled
16.2
15
PDB header: hydrolaseChain: A: PDB Molecule: beta-xylanase;PDBTitle: crystal structure of endo-1,4-beta-xylanase-like protein from2 acremonium chrysogenum
32 d1t3qa1
not modelled
15.9
12
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
33 c1knyA_
not modelled
15.6
16
PDB header: transferaseChain: A: PDB Molecule: kanamycin nucleotidyltransferase;PDBTitle: kanamycin nucleotidyltransferase
34 c6mbdD_
not modelled
15.4
28
PDB header: apoptosisChain: D: PDB Molecule: dm1;PDBTitle: human mcl-1 in complex with the designed peptide dm1
35 c6mbdC_
not modelled
15.4
28
PDB header: apoptosisChain: C: PDB Molecule: dm1;PDBTitle: human mcl-1 in complex with the designed peptide dm1
36 c4i7hA_
not modelled
15.2
7
PDB header: transcriptionChain: A: PDB Molecule: peroxide stress sensing regulator;PDBTitle: structural basis for peroxide sensing and gene regulation by perr from2 streptococcus pyogenes
37 c6fheA_
not modelled
15.0
8
PDB header: artificial enzymeChain: A: PDB Molecule: synthetic construct;PDBTitle: highly active enzymes by automated modular backbone assembly and2 sequence design
38 d1r85a_
not modelled
14.6
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
39 d1uh6a_
not modelled
14.5
23
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related
40 c6hulB_
not modelled
14.0
19
PDB header: lyaseChain: B: PDB Molecule: tryptophan synthase beta chain 1;PDBTitle: sulfolobus solfataricus tryptophan synthase ab complex
41 c1m94A_
not modelled
13.7
15
PDB header: structural genomics, signaling proteinChain: A: PDB Molecule: protein ynr032c-a;PDBTitle: solution structure of the yeast ubiquitin-like modifier2 protein hub1
42 d1m94a_
not modelled
13.7
15
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related
43 d1n62a1
not modelled
13.7
4
Fold: CO dehydrogenase ISP C-domain likeSuperfamily: CO dehydrogenase ISP C-domain likeFamily: CO dehydrogenase ISP C-domain like
44 c3wwnB_
not modelled
13.7
36
PDB header: metal binding protein/transferaseChain: B: PDB Molecule: orff;PDBTitle: crystal structure of lysz from thermus thermophilus complex with lysw
45 d1v6ya_
not modelled
13.2
5
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
46 c6ahtA_
not modelled
13.0
9
PDB header: transcriptionChain: A: PDB Molecule: conserved hypothetical plasmid protein;PDBTitle: plasmid partitioning protein tubr from bacillus cereus
47 d1jb0l_
not modelled
13.0
30
Fold: Photosystem I reaction center subunit XI, PsaLSuperfamily: Photosystem I reaction center subunit XI, PsaLFamily: Photosystem I reaction center subunit XI, PsaL
48 d1v6wa2
not modelled
12.9
5
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
49 c4upcA_
not modelled
12.7
20
PDB header: protein bindingChain: A: PDB Molecule: nicastrin;PDBTitle: structure of a extracellular domain
50 c3vpbF_
not modelled
12.7
40
PDB header: ligaseChain: F: PDB Molecule: alpha-aminoadipate carrier protein lysw;PDBTitle: argx from sulfolobus tokodaii complexed with2 lysw/glu/adp/mg/zn/sulfate
51 c3b9jI_
not modelled
12.7
4
PDB header: oxidoreductaseChain: I: PDB Molecule: xanthine oxidase;PDBTitle: structure of xanthine oxidase with 2-hydroxy-6-methylpurine
52 c3ibqA_
not modelled
12.6
19
PDB header: transferaseChain: A: PDB Molecule: pyridoxal kinase;PDBTitle: crystal structure of pyridoxal kinase from lactobacillus plantarum in2 complex with atp
53 d1tuxa_
not modelled
12.5
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
54 d1v0la_
not modelled
12.4
10
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
55 d1ylqa1
not modelled
12.4
30
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Catalytic subunit of bi-partite nucleotidyltransferase
56 d1bg4a_
not modelled
12.4
13
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
57 d1ta3b_
not modelled
12.3
18
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
58 c5k3jA_
not modelled
12.2
38
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coenzyme a oxidase;PDBTitle: crystals structure of acyl-coa oxidase-2 in caenorhabditis elegans2 bound with fad, ascaroside-coa, and atp
59 c4xx6A_
not modelled
12.2
8
PDB header: hydrolaseChain: A: PDB Molecule: beta-xylanase;PDBTitle: crystal structure of a glycosylated endo-beta-1,4-xylanase (glycoside2 hydrolase family 10/gh10) enzyme from gloeophyllum trabeum
60 d1exta3
not modelled
12.1
40
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
61 c3smpA_
not modelled
12.0
21
PDB header: transferaseChain: A: PDB Molecule: pantothenate kinase 1;PDBTitle: monoclinic crystal structure of human pantothenate kinase 1 alpha
62 c6fhfA_
not modelled
12.0
13
PDB header: artificial enzymeChain: A: PDB Molecule: design;PDBTitle: highly active enzymes by automated modular backbone assembly and2 sequence design
63 c4f8xA_
not modelled
11.6
8
PDB header: hydrolaseChain: A: PDB Molecule: endo-1,4-beta-xylanase;PDBTitle: penicillium canescens endo-1,4-beta-xylanase xyle
64 c1rm6F_
not modelled
11.6
12
PDB header: oxidoreductaseChain: F: PDB Molecule: 4-hydroxybenzoyl-coa reductase gamma subunit;PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
65 c5y6qA_
not modelled
11.5
19
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde oxidase small subunit;PDBTitle: crystal structure of an aldehyde oxidase from methylobacillus sp.2 ky4400
66 c3dwgA_
not modelled
11.4
8
PDB header: transferaseChain: A: PDB Molecule: cysteine synthase b;PDBTitle: crystal structure of a sulfur carrier protein complex found in the2 cysteine biosynthetic pathway of mycobacterium tuberculosis
67 c2rkbE_
not modelled
11.3
10
PDB header: lyaseChain: E: PDB Molecule: serine dehydratase-like;PDBTitle: serine dehydratase like-1 from human cancer cells
68 c2rffA_
not modelled
11.3
19
PDB header: transferaseChain: A: PDB Molecule: putative nucleotidyltransferase;PDBTitle: crystal structure of a putative nucleotidyltransferase (np_343093.1)2 from sulfolobus solfataricus at 1.40 a resolution
69 c5ys9A_
not modelled
11.2
38
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coenzyme a oxidase 3;PDBTitle: crystal structure of acyl-coa oxidase3 from yarrowia lipolytica
70 c3w26A_
not modelled
11.0
13
PDB header: hydrolaseChain: A: PDB Molecule: glycoside hydrolase family 10;PDBTitle: the high-resolution crystal structure of tsxyla, intracellular2 xylanase from /thermoanaerobacterium saccharolyticum jw/sl-ys485/:3 the complex of the e146a mutant with xylotriose
71 c5k3iH_
not modelled
10.9
44
PDB header: oxidoreductaseChain: H: PDB Molecule: acyl-coenzyme a oxidase;PDBTitle: crystal structure of acyl-coa oxidase-1 in caenorhabditis elegans2 complexed with fad and atp
72 d1h72c1
not modelled
10.9
33
Fold: Ribosomal protein S5 domain 2-likeSuperfamily: Ribosomal protein S5 domain 2-likeFamily: GHMP Kinase, N-terminal domain
73 c5nqgB_
not modelled
10.8
63
PDB header: cell invasionChain: B: PDB Molecule: ron2;PDBTitle: crystal structure of plasmodium vivax ama1 in complex with a 39 aa2 pvron2 peptide
74 c4qysA_
not modelled
10.8
14
PDB header: lyaseChain: A: PDB Molecule: tryptophan synthase beta chain 2;PDBTitle: trpb2 enzymes
75 d1knya2
not modelled
10.8
16
Fold: NucleotidyltransferaseSuperfamily: NucleotidyltransferaseFamily: Kanamycin nucleotidyltransferase (KNTase), N-terminal domain
76 c2fglA_
not modelled
10.5
13
PDB header: hydrolaseChain: A: PDB Molecule: alkaline thermostable endoxylanase;PDBTitle: an alkali thermostable f/10 xylanase from alkalophilic bacillus sp.2 ng-27
77 c1n60D_
not modelled
10.4
4
PDB header: oxidoreductaseChain: D: PDB Molecule: carbon monoxide dehydrogenase small chain;PDBTitle: crystal structure of the cu,mo-co dehydrogenase (codh); cyanide-2 inactivated form
78 c2k9iB_
not modelled
10.4
18
PDB header: dna binding proteinChain: B: PDB Molecule: uncharacterized protein orf56;PDBTitle: nmr structure of plasmid copy control protein orf56 from sulfolobus2 islandicus
79 c4pmyB_
not modelled
10.4
3
PDB header: hydrolaseChain: B: PDB Molecule: xylanase;PDBTitle: crystal structure of gh10 endo-b-1,4-xylanase (xynb) from xanthomonas2 axonopodis pv citri complexed with xylose
80 d1i1wa_
not modelled
10.4
15
Fold: TIM beta/alpha-barrelSuperfamily: (Trans)glycosidasesFamily: beta-glycanases
81 c3eubJ_
not modelled
10.4
4
PDB header: oxidoreductaseChain: J: PDB Molecule: xanthine dehydrogenase/oxidase;PDBTitle: crystal structure of desulfo-xanthine oxidase with xanthine
82 c6ju2A_
not modelled
10.3
35
PDB header: viral protein/rnaChain: A: PDB Molecule: polymerase 3;PDBTitle: structure of influenza d virus polymerase bound to crna promoter in2 class 1
83 d1p0ra_
not modelled
10.0
26
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related
84 d1rypa_
not modelled
10.0
11
Fold: Ntn hydrolase-likeSuperfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)Family: Proteasome subunits
85 c1ffuA_
not modelled
9.9
12
PDB header: hydrolaseChain: A: PDB Molecule: cuts, iron-sulfur protein of carbon monoxidePDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava which lacks the mo-pyranopterin moiety of the3 molybdenum cofactor
86 c2lr8A_
not modelled
9.9
16
PDB header: apoptosisChain: A: PDB Molecule: casp8-associated protein 2;PDBTitle: solution nmr structure of casp8-associated protein 2 from homo2 sapiens, northeast structural genomics consortium (nesg) target3 hr8150a
87 c5y9dA_
not modelled
9.8
40
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coenzyme a oxidase 1;PDBTitle: crystal structure of acyl-coa oxidase1 from yarrowia lipolytica
88 c2ddhA_
not modelled
9.8
54
PDB header: oxidoreductaseChain: A: PDB Molecule: acyl-coa oxidase;PDBTitle: crystal structure of acyl-coa oxidase complexed with 3-oh-dodecanoate
89 c4hx3H_
not modelled
9.7
50
PDB header: hydrolase/hydrolase inhibitorChain: H: PDB Molecule: neutral proteinase inhibitor scnpi;PDBTitle: crystal structure of streptomyces caespitosus sermetstatin in complex2 with s. caespitosus snapalysin
90 c5m0kB_
not modelled
9.6
10
PDB header: hydrolaseChain: B: PDB Molecule: beta-xylanase;PDBTitle: crystal structure of endo-1,4-beta-xylanase from cellulomonas2 flavigena
91 c5oeiA_
not modelled
9.5
16
PDB header: transport proteinChain: A: PDB Molecule: uncharacterized protein family upf0065:tat pathway signal;PDBTitle: r. palustris rpa4515 with oxoadipate
92 c2mpoA_
not modelled
9.4
67
PDB header: cell adhesionChain: A: PDB Molecule: mic2-associated protein;PDBTitle: structural basis of toxoplasma gondii mic2-associated protein2 interaction with mic2
93 c1us2A_
not modelled
9.3
10
PDB header: hydrolaseChain: A: PDB Molecule: endo-beta-1,4-xylanase;PDBTitle: xylanase10c (mutant e385a) from cellvibrio japonicus in complex with2 xylopentaose
94 d1pbya1
not modelled
9.2
14
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Quinohemoprotein amine dehydrogenase A chain, domains 1 and 2
95 c2zsjB_
not modelled
9.2
25
PDB header: lyaseChain: B: PDB Molecule: threonine synthase;PDBTitle: crystal structure of threonine synthase from aquifex aeolicus vf5
96 c5tzbA_
not modelled
9.2
15
PDB header: hydrolaseChain: A: PDB Molecule: d-aminopeptidase;PDBTitle: burkholderia sp. beta-aminopeptidase
97 c6q8nB_
not modelled
9.1
10
PDB header: hydrolaseChain: B: PDB Molecule: beta-xylanase;PDBTitle: gh10 endo-xylanase in complex with xylobiose epoxide inhibitor
98 c3hrdH_
not modelled
9.1
12
PDB header: oxidoreductaseChain: H: PDB Molecule: nicotinate dehydrogenase small fes subunit;PDBTitle: crystal structure of nicotinate dehydrogenase
99 d2nrha1
not modelled
9.1
6
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: CoaX-like