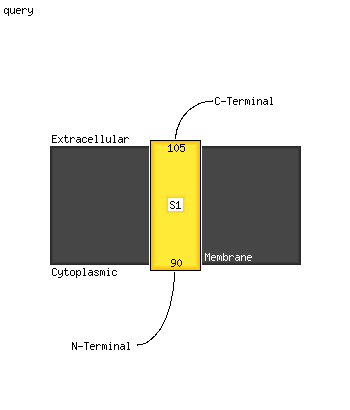
1 c3dzzB_
100.0
25
PDB header: transferaseChain: B: PDB Molecule: putative pyridoxal 5'-phosphate-dependent c-s lyase;PDBTitle: crystal structure of a putative plp-dependent aminotransferase2 (lbul_1103) from lactobacillus delbrueckii subsp. at 1.61 a3 resolution
2 c3b1dD_
100.0
23
PDB header: lyaseChain: D: PDB Molecule: betac-s lyase;PDBTitle: crystal structure of betac-s lyase from streptococcus anginosus in2 complex with l-serine: external aldimine form
3 c3fdbA_
100.0
41
PDB header: transferaseChain: A: PDB Molecule: putative plp-dependent beta-cystathionase;PDBTitle: crystal structure of a putative plp-dependent beta-cystathionase2 (aecd, dip1736) from corynebacterium diphtheriae at 1.99 a resolution
4 d1c7na_
100.0
24
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like
5 c4dgtA_
100.0
31
PDB header: transferaseChain: A: PDB Molecule: putative pyridoxal phosphate-dependent transferase;PDBTitle: crystal structure of plp-bound putative aminotransferase from2 clostridium difficile 630 crystallized with magnesium formate
6 c3tcmB_
100.0
16
PDB header: transferaseChain: B: PDB Molecule: alanine aminotransferase 2;PDBTitle: crystal structure of alanine aminotransferase from hordeum vulgare
7 c3l8aB_
100.0
22
PDB header: lyaseChain: B: PDB Molecule: putative aminotransferase, probable beta-cystathionase;PDBTitle: crystal structure of metc from streptococcus mutans
8 c5z0qG_
100.0
31
PDB header: transferaseChain: G: PDB Molecule: aminotransferase, class i and ii;PDBTitle: crystal structure of ovob
9 c3kaxB_
100.0
23
PDB header: lyaseChain: B: PDB Molecule: aminotransferase, classes i and ii;PDBTitle: crystal structure of a putative c-s lyase from bacillus anthracis
10 c5yhvA_
100.0
21
PDB header: transferaseChain: A: PDB Molecule: aminotransferase;PDBTitle: crystal structure of an aminotransferase from mycobacterium2 tuberculosis
11 c3g0tA_
100.0
13
PDB header: transferaseChain: A: PDB Molecule: putative aminotransferase;PDBTitle: crystal structure of putative aspartate aminotransferase (np_905498.1)2 from porphyromonas gingivalis w83 at 1.75 a resolution
12 c3h14A_
100.0
21
PDB header: transferaseChain: A: PDB Molecule: aminotransferase, classes i and ii;PDBTitle: crystal structure of a putative aminotransferase from silicibacter2 pomeroyi
13 c3b46B_
100.0
18
PDB header: transferaseChain: B: PDB Molecule: aminotransferase bna3;PDBTitle: crystal structure of bna3p, a putative kynurenine2 aminotransferase from saccharomyces cerevisiae
14 c3e2yB_
100.0
22
PDB header: transferase, lyaseChain: B: PDB Molecule: kynurenine-oxoglutarate transaminase 3;PDBTitle: crystal structure of mouse kynurenine aminotransferase iii in complex2 with glutamine
15 d1u08a_
100.0
21
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
16 d2r5ea1
100.0
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
17 d1xi9a_
100.0
22
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
18 d1j32a_
100.0
23
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
19 d1w7la_
100.0
18
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
20 c1d2fB_
100.0
27
PDB header: transferaseChain: B: PDB Molecule: maly protein;PDBTitle: x-ray structure of maly from escherichia coli: a pyridoxal-5'-2 phosphate-dependent enzyme acting as a modulator in mal gene3 expression
21 c3qguB_
not modelled
100.0
18
PDB header: transferaseChain: B: PDB Molecule: ll-diaminopimelate aminotransferase;PDBTitle: l,l-diaminopimelate aminotransferase from chalmydomonas reinhardtii
22 c4my5C_
not modelled
100.0
20
PDB header: transferaseChain: C: PDB Molecule: putative amino acid aminotransferase;PDBTitle: crystal structure of the aromatic amino acid aminotransferase from2 streptococcus mutants
23 c2o0rA_
not modelled
100.0
26
PDB header: transferaseChain: A: PDB Molecule: rv0858c (n-succinyldiaminopimelate aminotransferase);PDBTitle: the three-dimensional structure of n-succinyldiaminopimelate2 aminotransferase from mycobacterium tuberculosis
24 d1d2fa_
not modelled
100.0
27
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Cystathionine synthase-like
25 c3eibB_
not modelled
100.0
19
PDB header: transferaseChain: B: PDB Molecule: ll-diaminopimelate aminotransferase;PDBTitle: crystal structure of k270n variant of ll-diaminopimelate2 aminotransferase from arabidopsis thaliana
26 d1o4sa_
not modelled
100.0
24
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
27 c3if2B_
not modelled
100.0
18
PDB header: transferaseChain: B: PDB Molecule: aminotransferase;PDBTitle: crystal structure of putative amino-acid aminotransferase2 (yp_265399.1) from psychrobacter arcticum 273-4 at 2.50 a resolution
28 c2o1bA_
not modelled
100.0
18
PDB header: transferaseChain: A: PDB Molecule: aminotransferase, class i;PDBTitle: structure of aminotransferase from staphylococcus aureus
29 c6f35B_
not modelled
100.0
20
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase b;PDBTitle: crystal structure of the aspartate aminotranferase from rhizobium2 meliloti
30 c2zc0C_
not modelled
100.0
16
PDB header: transferaseChain: C: PDB Molecule: alanine glyoxylate transaminase;PDBTitle: crystal structure of an archaeal alanine:glyoxylate aminotransferase
31 c3jtxB_
not modelled
100.0
20
PDB header: transferaseChain: B: PDB Molecule: aminotransferase;PDBTitle: crystal structure of aminotransferase (np_283882.1) from neisseria2 meningitidis z2491 at 1.91 a resolution
32 c3ihjA_
not modelled
100.0
17
PDB header: transferaseChain: A: PDB Molecule: alanine aminotransferase 2;PDBTitle: human alanine aminotransferase 2 in complex with plp
33 c1ynuA_
not modelled
100.0
19
PDB header: lyaseChain: A: PDB Molecule: 1-aminocyclopropane-1-carboxylate synthase;PDBTitle: crystal structure of apple acc synthase in complex with l-vinylglycine
34 c6f77D_
not modelled
100.0
22
PDB header: transferaseChain: D: PDB Molecule: aspartate aminotransferase a;PDBTitle: crystal structure of the prephenate aminotransferase from rhizobium2 meliloti
35 c5wmiA_
not modelled
100.0
23
PDB header: transferaseChain: A: PDB Molecule: bifunctional aspartate aminotransferase andPDBTitle: arabidopsis thaliana prephenate aminotransferase mutant- t84v
36 d1bw0a_
not modelled
100.0
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
37 c3nraA_
not modelled
100.0
20
PDB header: transferaseChain: A: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of an aspartate aminotransferase (yp_354942.1) from2 rhodobacter sphaeroides 2.4.1 at 2.15 a resolution
38 c4fl0A_
not modelled
100.0
17
PDB header: transferaseChain: A: PDB Molecule: aminotransferase ald1;PDBTitle: crystal structure of ald1 from arabidopsis thaliana
39 d1b5pa_
not modelled
100.0
22
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
40 d1gdea_
not modelled
100.0
23
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
41 c2x5dD_
not modelled
100.0
25
PDB header: transferaseChain: D: PDB Molecule: probable aminotransferase;PDBTitle: crystal structure of a probable aminotransferase from pseudomonas2 aeruginosa
42 d2gb3a1
not modelled
100.0
18
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
43 c4cvqB_
not modelled
100.0
18
PDB header: transferaseChain: B: PDB Molecule: glutamate-pyruvate aminotransferase alaa;PDBTitle: crystal structure of an aminotransferase from escherichia coli at 2.2 11 angstroem resolution
44 c3eleB_
not modelled
100.0
20
PDB header: transferaseChain: B: PDB Molecule: amino transferase;PDBTitle: crystal structure of amino transferase (rer070207001803) from2 eubacterium rectale at 2.10 a resolution
45 d1iaya_
not modelled
100.0
20
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
46 c3pplB_
not modelled
100.0
18
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of an aspartate transaminase (ncgl0237, cgl0240)2 from corynebacterium glutamicum atcc 13032 kitasato at 1.25 a3 resolution
47 d1wsta1
not modelled
100.0
18
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
48 d1vp4a_
not modelled
100.0
17
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
49 c3ezsB_
not modelled
100.0
16
PDB header: transferaseChain: B: PDB Molecule: aminotransferase aspb;PDBTitle: crystal structure of aminotransferase aspb (np_207418.1) from2 helicobacter pylori 26695 at 2.19 a resolution
50 d1m7ya_
not modelled
100.0
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
51 c6hnuA_
not modelled
100.0
19
PDB header: transferaseChain: A: PDB Molecule: aromatic amino acid aminotransferase i;PDBTitle: crystal structure of the aminotransferase aro8 from c. albicans with2 ligands
52 c4je5C_
not modelled
100.0
18
PDB header: transferaseChain: C: PDB Molecule: aromatic/aminoadipate aminotransferase 1;PDBTitle: crystal structure of the aromatic aminotransferase aro8, a putative2 alpha-aminoadipate aminotransferase in saccharomyces cerevisiae
53 c3asaA_
not modelled
100.0
21
PDB header: transferaseChain: A: PDB Molecule: ll-diaminopimelate aminotransferase;PDBTitle: crystal structure of apo-ll-diaminopimelate aminotransferase from2 chlamydia trachomatis
54 c2douA_
not modelled
100.0
24
PDB header: transferaseChain: A: PDB Molecule: probable n-succinyldiaminopimelate aminotransferase;PDBTitle: probable n-succinyldiaminopimelate aminotransferase (ttha0342) from2 thermus thermophilus hb8
55 d1x0ma1
not modelled
100.0
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
56 c6hndA_
not modelled
100.0
15
PDB header: transferaseChain: A: PDB Molecule: aromatic-amino-acid:2-oxoglutarate transaminase;PDBTitle: crystal structure of the aromatic aminotransferase aro9 from c.2 albicans
57 c2x5fB_
not modelled
100.0
14
PDB header: transferaseChain: B: PDB Molecule: aspartate_tyrosine_phenylalanine pyridoxal-5'PDBTitle: crystal structure of the methicillin-resistant2 staphylococcus aureus sar2028, an3 aspartate_tyrosine_phenylalanine pyridoxal-5'-phosphate4 dependent aminotransferase
58 c4ix8B_
not modelled
100.0
22
PDB header: transferaseChain: B: PDB Molecule: tyrosine aminotransferase;PDBTitle: crystal structure of tyrosine aminotransferase from leishmania2 infantum
59 c5c6uA_
not modelled
100.0
14
PDB header: transferaseChain: A: PDB Molecule: aminotransferase;PDBTitle: rv3722c aminotransferase from mycobacterium tuberculosis
60 c3pdxA_
not modelled
100.0
18
PDB header: transferaseChain: A: PDB Molecule: tyrosine aminotransferase;PDBTitle: crystal structural of mouse tyrosine aminotransferase
61 c3t18D_
not modelled
100.0
14
PDB header: transferaseChain: D: PDB Molecule: aminotransferase class i and ii;PDBTitle: crystal structure of aminotransferase from anaerococcus prevotii dsm2 20548.
62 c3dydB_
not modelled
100.0
19
PDB header: transferaseChain: B: PDB Molecule: tyrosine aminotransferase;PDBTitle: human tyrosine aminotransferase
63 c3g7qA_
not modelled
100.0
17
PDB header: transferaseChain: A: PDB Molecule: valine-pyruvate aminotransferase;PDBTitle: crystal structure of valine-pyruvate aminotransferase avta2 (np_462565.1) from salmonella typhimurium lt2 at 1.80 a resolution
64 c3dc1A_
not modelled
100.0
17
PDB header: transferaseChain: A: PDB Molecule: kynurenine/alpha-aminoadipate aminotransferasePDBTitle: crystal structure of kynurenine aminotransferase ii complex with2 alpha-ketoglutarate
65 c3ez1A_
not modelled
100.0
15
PDB header: transferaseChain: A: PDB Molecule: aminotransferase mocr family;PDBTitle: crystal structure of putative aminotransferase (mocr family)2 (yp_604413.1) from deinococcus geothermalis dsm 11300 at 2.60 a3 resolution
66 c2z61A_
not modelled
100.0
22
PDB header: transferaseChain: A: PDB Molecule: probable aspartate aminotransferase 2;PDBTitle: crystal structure of mj0684 from methanococcus jannaschii2 reveals its similarity in the active site to kynurenine3 aminotransferases
67 c4n0bA_
not modelled
100.0
14
PDB header: transcription activatorChain: A: PDB Molecule: hth-type transcriptional regulatory protein gabr;PDBTitle: crystal structure of bacillus subtilis gabr, an autorepressor and2 transcriptional activator of gabt
68 c6c3aB_
not modelled
100.0
15
PDB header: biosynthetic proteinChain: B: PDB Molecule: uncharacterized protein;PDBTitle: o2-, plp-dependent l-arginine hydroxylase rohp 4-hydroxy-2-2 ketoarginine complex
69 c3op7A_
not modelled
100.0
20
PDB header: transferaseChain: A: PDB Molecule: aminotransferase class i and ii;PDBTitle: crystal structure of a plp-dependent aminotransferase (zp_03625122.1)2 from streptococcus suis 89-1591 at 1.70 a resolution
70 d2csta_
not modelled
100.0
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
71 d1v2da_
not modelled
100.0
23
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
72 c4rkdA_
not modelled
100.0
15
PDB header: transferaseChain: A: PDB Molecule: aromatic amino acid aminotransferase;PDBTitle: psychrophilic aromatic amino acids aminotransferase from psychrobacter2 sp. b6 cocrystalized with aspartic acid
73 c2zy4F_
not modelled
100.0
14
PDB header: lyaseChain: F: PDB Molecule: l-aspartate beta-decarboxylase;PDBTitle: dodecameric l-aspartate beta-decarboxylase
74 c4wd2A_
not modelled
100.0
17
PDB header: transferaseChain: A: PDB Molecule: aromatic-amino-acid transaminase tyrb;PDBTitle: crystal structure of an aromatic amino acid aminotransferase from2 burkholderia cenocepacia j2315
75 c3k7yA_
not modelled
100.0
16
PDB header: transferaseChain: A: PDB Molecule: aspartate aminotransferase;PDBTitle: aspartate aminotransferase of plasmodium falciparum
76 c3rq1A_
not modelled
100.0
15
PDB header: transferaseChain: A: PDB Molecule: aminotransferase class i and ii;PDBTitle: crystal structure of aminotransferase class i and ii from veillonella2 parvula
77 c3cbfA_
not modelled
100.0
16
PDB header: transferaseChain: A: PDB Molecule: alpha-aminodipate aminotransferase;PDBTitle: crystal structure of lysn, alpha-aminoadipate2 aminotransferase, from thermus thermophilus hb27
78 d1ajsa_
not modelled
100.0
18
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
79 c4effA_
not modelled
100.0
22
PDB header: transferaseChain: A: PDB Molecule: aromatic-amino-acid aminotransferase;PDBTitle: crystal structure of aromatic-amino-acid aminotransferase from2 burkholderia pseudomallei
80 c2zy3A_
not modelled
100.0
16
PDB header: lyaseChain: A: PDB Molecule: l-aspartate beta-decarboxylase;PDBTitle: dodecameric l-aspartate beta-decarboxylase
81 d2q7wa1
not modelled
100.0
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
82 c4eu1A_
not modelled
100.0
16
PDB header: transferaseChain: A: PDB Molecule: mitochondrial aspartate aminotransferase;PDBTitle: structure of a mitochondrial aspartate aminotransferase from2 trypanosoma brucei
83 d2ay1a_
not modelled
100.0
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
84 c3f6tA_
not modelled
100.0
14
PDB header: transferaseChain: A: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of aspartate aminotransferase (e.c. 2.6.1.1)2 (yp_194538.1) from lactobacillus acidophilus ncfm at 2.15 a3 resolution
85 d7aata_
not modelled
100.0
14
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
86 c4h51B_
not modelled
100.0
16
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of a putative aspartate aminotransferase from2 leishmania major friedlin
87 c3ly1C_
not modelled
100.0
15
PDB header: transferaseChain: C: PDB Molecule: putative histidinol-phosphate aminotransferase;PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (yp_050345.1) from erwinia carotovora atroseptica scri1043 at 1.80 a3 resolution
88 d1yaaa_
not modelled
100.0
18
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
89 c3d6kB_
not modelled
100.0
17
PDB header: transferaseChain: B: PDB Molecule: putative aminotransferase;PDBTitle: the crystal structure of a putative aminotransferase from2 corynebacterium diphtheriae
90 c4m2jA_
not modelled
100.0
22
PDB header: transferaseChain: A: PDB Molecule: aminotransferase;PDBTitle: crystal structure of plp-dependent cyclase orfr in complex with au
91 c4wbtA_
not modelled
100.0
18
PDB header: transferaseChain: A: PDB Molecule: probable histidinol-phosphate aminotransferase;PDBTitle: crystal structure of histidinol-phosphate aminotransferase from2 sinorhizobium meliloti in complex with pyridoxal-5'-phosphate
92 c4r8dB_
not modelled
100.0
18
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of rv1600 encoded aminotransferase in complex with2 plp-mes from mycobacterium tuberculosis
93 c3bwnF_
not modelled
100.0
16
PDB header: transferaseChain: F: PDB Molecule: l-tryptophan aminotransferase;PDBTitle: l-tryptophan aminotransferase
94 d3tata_
not modelled
100.0
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
95 c3cq6E_
not modelled
100.0
16
PDB header: transferaseChain: E: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: histidinol-phosphate aminotransferase from corynebacterium2 glutamicum holo-form (plp covalently bound )
96 c3mebB_
not modelled
100.0
19
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase;PDBTitle: structure of cytoplasmic aspartate aminotransferase from giardia2 lamblia
97 d1lc5a_
not modelled
100.0
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
98 c3getA_
not modelled
100.0
14
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of putative histidinol-phosphate aminotransferase2 (np_281508.1) from campylobacter jejuni at 2.01 a resolution
99 c4r2nA_
not modelled
100.0
19
PDB header: transferaseChain: A: PDB Molecule: putative phenylalanine aminotransferase;PDBTitle: crystal structure of rv3772 in complex with its substrate
100 c6ezlB_
not modelled
100.0
16
PDB header: transferaseChain: B: PDB Molecule: aspartate aminotransferase;PDBTitle: crystal structure of aspartate aminotransferase from trypanosoma cruzi2 at 2.07 angstrom resolution
101 d2hoxa1
not modelled
100.0
15
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
102 c5dj3B_
not modelled
100.0
19
PDB header: transferaseChain: B: PDB Molecule: plp-dependent l-arginine hydroxylase mppp;PDBTitle: structure of the plp-dependent l-arginine hydroxylase mppp with d-2 arginine bound
103 c3eucB_
not modelled
100.0
17
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase 2;PDBTitle: crystal structure of histidinol-phosphate aminotransferase2 (yp_297314.1) from ralstonia eutropha jmp134 at 2.05 a resolution
104 c3hdoB_
not modelled
100.0
18
PDB header: transferaseChain: B: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: crystal structure of a histidinol-phosphate aminotransferase from2 geobacter metallireducens
105 d1fg7a_
not modelled
100.0
19
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
106 c3ftbA_
not modelled
100.0
13
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: the crystal structure of the histidinol-phosphate2 aminotransferase from clostridium acetobutylicum
107 d2f8ja1
not modelled
100.0
20
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: AAT-like
108 c3fkdC_
not modelled
100.0
14
PDB header: lyaseChain: C: PDB Molecule: l-threonine-o-3-phosphate decarboxylase;PDBTitle: the crystal structure of l-threonine-o-3-phosphate decarboxylase from2 porphyromonas gingivalis
109 c3ffhA_
not modelled
100.0
17
PDB header: transferaseChain: A: PDB Molecule: histidinol-phosphate aminotransferase;PDBTitle: the crystal structure of histidinol-phosphate aminotransferase from2 listeria innocua clip11262.
110 c3p1tB_
not modelled
100.0
20
PDB header: transferaseChain: B: PDB Molecule: putative histidinol-phosphate aminotransferase;PDBTitle: crystal structure of a putative aminotransferase (bpsl1724) from2 burkholderia pseudomallei k96243 at 2.60 a resolution
111 c6ouxB_
not modelled
100.0
14
PDB header: lyaseChain: B: PDB Molecule: threonine phosphate decarboxylase-like enzyme;PDBTitle: structure of smul_1544, a decarboxylase from sulfurospirillum2 multivorans
112 c2w8wA_
not modelled
100.0
15
PDB header: transferaseChain: A: PDB Molecule: serine palmitoyltransferase;PDBTitle: n100y spt with plp-ser
113 c3a2bA_
not modelled
100.0
13
PDB header: transferaseChain: A: PDB Molecule: serine palmitoyltransferase;PDBTitle: crystal structure of serine palmitoyltransferase from sphingobacterium2 multivorum with substrate l-serine
114 c5jayB_
not modelled
100.0
14
PDB header: transferaseChain: B: PDB Molecule: 8-amino-7-oxononanoate synthase;PDBTitle: crystal structure of an 8-amino-7-oxononanoate synthase from2 burkholderia xenovorans
115 c3wy7D_
not modelled
100.0
17
PDB header: transferaseChain: D: PDB Molecule: 8-amino-7-oxononanoate synthase;PDBTitle: crystal structure of mycobacterium smegmatis 7-keto-8-aminopelargonic2 acid (kapa) synthase biof
116 c3hqtB_
not modelled
100.0
14
PDB header: transferaseChain: B: PDB Molecule: cai-1 autoinducer synthase;PDBTitle: plp-dependent acyl-coa transferase cqsa
117 c2ordA_
not modelled
100.0
14
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
118 c3nx3A_
not modelled
100.0
9
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase (argd) from2 campylobacter jejuni
119 c4addD_
not modelled
100.0
14
PDB header: transferaseChain: D: PDB Molecule: succinylornithine transaminase;PDBTitle: structural and functional study of succinyl-ornithine transaminase2 from e. coli
120 c4uoxB_
not modelled
100.0
12
PDB header: transferaseChain: B: PDB Molecule: putrescine aminotransferase;PDBTitle: crystal structure of ygjg in complex with pyridoxal-5'-phosphate2 and putrescine