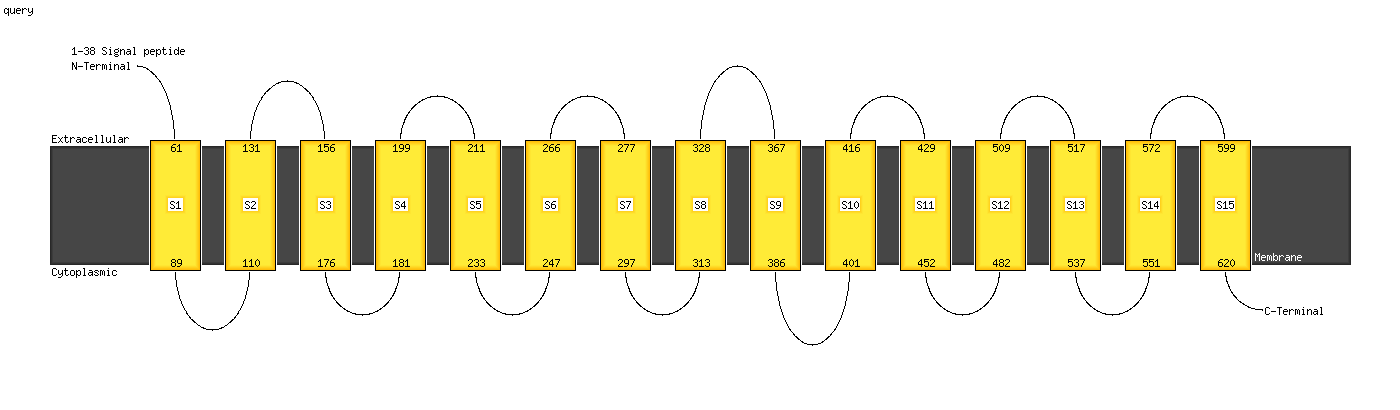
1 c4flaC_
44.9
31
PDB header: transcriptionChain: C: PDB Molecule: regulation of nuclear pre-mrna domain-containing proteinPDBTitle: crystal structure of human rprd1b, carboxy-terminal domain
2 c5fwlE_
41.7
19
PDB header: chaperoneChain: E: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: atomic cryoem structure of hsp90-cdc37-cdk4 complex
3 c5kxfA_
41.0
29
PDB header: rna binding proteinChain: A: PDB Molecule: flowering time control protein fpa;PDBTitle: crystal structure of the spoc domain of the arabidopsis flowering2 regulator fpa
4 c5n9mA_
34.1
9
PDB header: transferaseChain: A: PDB Molecule: cobyric acid synthase;PDBTitle: crystal structure of gatd - a glutamine amidotransferase from2 staphylococcus aureus involved in peptidoglycan amidation
5 c2w8aC_
32.0
7
PDB header: membrane proteinChain: C: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of the sodium-coupled glycine betaine2 symporter betp from corynebacterium glutamicum with bound3 substrate
6 d1v4pa_
31.2
21
Fold: RRF/tRNA synthetase additional domain-likeSuperfamily: ThrRS/AlaRS common domainFamily: AlaX-like
7 c6bn1B_
30.8
46
PDB header: signaling proteinChain: B: PDB Molecule: scaffold protein salvador;PDBTitle: salvador hippo sarah domain complex
8 c6ao5B_
30.8
38
PDB header: signaling proteinChain: B: PDB Molecule: protein salvador homolog 1;PDBTitle: crystal structure of human mst2 in complex with sav1 sarah domain
9 d1f6ga_
28.1
15
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels
10 c4ainB_
27.4
9
PDB header: membrane proteinChain: B: PDB Molecule: glycine betaine transporter betp;PDBTitle: crystal structure of betp with asymmetric protomers.
11 c2jo1A_
25.3
23
PDB header: hydrolase regulatorChain: A: PDB Molecule: phospholemman;PDBTitle: structure of the na,k-atpase regulatory protein fxyd1 in2 micelles
12 c6hwhX_
22.1
42
PDB header: electron transportChain: X: PDB Molecule: cytochrome c oxidase polypeptide 4;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
13 c1ddxA_
21.3
18
PDB header: oxidoreductaseChain: A: PDB Molecule: protein (prostaglandin h2 synthase-2);PDBTitle: crystal structure of a mixture of arachidonic acid and prostaglandin2 bound to the cyclooxygenase active site of cox-2: prostaglandin3 structure
14 c5z51A_
20.6
18
PDB header: transferaseChain: A: PDB Molecule: dna primase;PDBTitle: helicase binding domain of primase from mycobacterium tuberculosis
15 c3pghD_
20.6
18
PDB header: oxidoreductaseChain: D: PDB Molecule: cyclooxygenase-2;PDBTitle: cyclooxygenase-2 (prostaglandin synthase-2) complexed with a non-2 selective inhibitor, flurbiprofen
16 d1cvua1
20.6
18
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Myeloperoxidase-like
17 d1gvna_
18.1
33
Fold: immunoglobulin/albumin-binding domain-likeSuperfamily: Plasmid maintenance system epsilon/zeta, antidote epsilon subunitFamily: Plasmid maintenance system epsilon/zeta, antidote epsilon subunit
18 d1q4ga1
17.5
12
Fold: Heme-dependent peroxidasesSuperfamily: Heme-dependent peroxidasesFamily: Myeloperoxidase-like
19 c6hc2L_
14.5
25
PDB header: cell cycleChain: L: PDB Molecule: nuclear mitotic apparatus protein 1;PDBTitle: crystal structure of numa/lgn hetero-hexamers
20 d1q90d_
13.7
17
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
21 c4tq0D_
not modelled
13.0
24
PDB header: protein bindingChain: D: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg5-atg16n69
22 c4tq0B_
not modelled
12.6
24
PDB header: protein bindingChain: B: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg5-atg16n69
23 c2ksfA_
not modelled
11.0
21
PDB header: transferaseChain: A: PDB Molecule: sensor protein kdpd;PDBTitle: backbone structure of the membrane domain of e. coli histidine kinase2 receptor kdpd, center for structures of membrane proteins (csmp)3 target 4312c
24 c2lonA_
not modelled
10.9
21
PDB header: membrane proteinChain: A: PDB Molecule: hig1 domain family member 1b;PDBTitle: backbone structure of human membrane protein higd1b
25 d2vcha1
not modelled
10.7
5
Fold: UDP-Glycosyltransferase/glycogen phosphorylaseSuperfamily: UDP-Glycosyltransferase/glycogen phosphorylaseFamily: UDPGT-like
26 c5npwF_
not modelled
10.3
24
PDB header: cell cycleChain: F: PDB Molecule: autophagy-related protein 16-1;PDBTitle: structure of human atg5-atg16l1(atg5bd) complex (c2)
27 c5npvD_
not modelled
10.3
24
PDB header: cell cycleChain: D: PDB Molecule: autophagy-related protein 16-1;PDBTitle: structure of human atg5-atg16l1(atg5bd) complex (i4)
28 c2lomA_
not modelled
10.3
15
PDB header: membrane proteinChain: A: PDB Molecule: hig1 domain family member 1a;PDBTitle: backbone structure of human membrane protein higd1a
29 c5npvB_
not modelled
10.2
24
PDB header: cell cycleChain: B: PDB Molecule: autophagy-related protein 16-1;PDBTitle: structure of human atg5-atg16l1(atg5bd) complex (i4)
30 c3c4rC_
not modelled
9.8
21
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of an uncharacterized protein encoded by2 cryptic prophage
31 c2n28A_
not modelled
9.8
18
PDB header: viral proteinChain: A: PDB Molecule: protein vpu;PDBTitle: solid-state nmr structure of vpu
32 c2oyuP_
not modelled
9.6
12
PDB header: oxidoreductaseChain: P: PDB Molecule: prostaglandin g/h synthase 1;PDBTitle: indomethacin-(s)-alpha-ethyl-ethanolamide bound to cyclooxygenase-1
33 c3aqgB_
not modelled
9.5
50
PDB header: unknown functionChain: B: PDB Molecule: zymogen granule protein 16 homolog b;PDBTitle: crystal structure of human pancreatic secretory protein zg16b
34 c1ht8B_
not modelled
9.3
12
PDB header: oxidoreductaseChain: B: PDB Molecule: prostaglandin h2 synthase-1;PDBTitle: the 2.7 angstrom resolution model of ovine cox-1 complexed with2 alclofenac
35 c1pggB_
not modelled
9.3
12
PDB header: oxidoreductaseChain: B: PDB Molecule: prostaglandin h2 synthase-1;PDBTitle: prostaglandin h2 synthase-1 complexed with 1-(4-iodobenzoyl)-5-2 methoxy-2-methylindole-3-acetic acid (iodoindomethacin), trans model
36 c5d3aA_
not modelled
9.2
13
PDB header: motor proteinChain: A: PDB Molecule: kinesin-like protein kif21a;PDBTitle: kif21a regulatory coiled coil
37 c5zt3A_
not modelled
8.9
19
PDB header: plant proteinChain: A: PDB Molecule: wa352;PDBTitle: crystal structure of wa352 from oryza sativa
38 c3m6cA_
not modelled
8.3
18
PDB header: chaperoneChain: A: PDB Molecule: 60 kda chaperonin 1;PDBTitle: crystal structure of mycobacterium tuberculosis groel1 apical domain
39 d2e74b1
not modelled
8.3
14
Fold: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Superfamily: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)Family: a domain/subunit of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase)
40 c5whaF_
not modelled
8.2
20
PDB header: protein bindingChain: F: PDB Molecule: miniprotein 225-11;PDBTitle: kras g12v, bound to gdp and miniprotein 225-11
41 d1sjpa2
not modelled
8.0
12
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
42 c2rddB_
not modelled
7.6
13
PDB header: membrane protein/transport proteinChain: B: PDB Molecule: upf0092 membrane protein yajc;PDBTitle: x-ray crystal structure of acrb in complex with a novel2 transmembrane helix.
43 c3rgbG_
not modelled
7.5
20
PDB header: oxidoreductaseChain: G: PDB Molecule: methane monooxygenase subunit c2;PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
44 c4tt1A_
not modelled
7.4
21
PDB header: hydrolaseChain: A: PDB Molecule: deneddylase;PDBTitle: crystal structure of fragment 1600-1733 of hsv1 ul36, native
45 c2ncaA_
not modelled
7.3
21
PDB header: chaperoneChain: A: PDB Molecule: hsp90 co-chaperone cdc37;PDBTitle: structural model for the n-terminal domain of human cdc37
46 c4a1aI_
not modelled
7.3
17
PDB header: ribosomeChain: I: PDB Molecule: 60s ribosomal protein l13a;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 3.
47 c3j3bO_
not modelled
7.3
9
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l13a;PDBTitle: structure of the human 60s ribosomal proteins
48 c4r36A_
not modelled
7.1
12
PDB header: transferaseChain: A: PDB Molecule: putative acyl-[acyl-carrier-protein]--udp-n-PDBTitle: crystal structure analysis of lpxa, a udp-n-acetylglucosamine2 acyltransferase from bacteroides fragilis 9343
49 c2mbhB_
not modelled
7.1
17
PDB header: transcriptionChain: B: PDB Molecule: krueppel-like factor 1;PDBTitle: nmr structure of eklf(22-40)/ubiquitin complex
50 c3mawB_
not modelled
7.1
54
PDB header: viral proteinChain: B: PDB Molecule: fusion glycoprotein f0;PDBTitle: structure of the newcastle disease virus f protein in the post-fusion2 conformation
51 c4nawG_
not modelled
7.1
25
PDB header: protein transport/ligaseChain: G: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5-atg16n in complex with a2 fragment of atg3
52 c4nawO_
not modelled
7.1
25
PDB header: protein transport/ligaseChain: O: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5-atg16n in complex with a2 fragment of atg3
53 c5cdjA_
not modelled
7.0
10
PDB header: chaperoneChain: A: PDB Molecule: rubisco large subunit-binding protein subunit alpha,PDBTitle: apical domain of chloroplast chaperonin 60a
54 c4gdkC_
not modelled
6.9
25
PDB header: protein bindingChain: C: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5 conjugate in complex with an n-2 terminal fragment of atg16l1
55 c4gdlC_
not modelled
6.9
25
PDB header: protein bindingChain: C: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5 conjugate in complex with an n-2 terminal fragment of atg16l1
56 c4nawC_
not modelled
6.9
25
PDB header: protein transport/ligaseChain: C: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5-atg16n in complex with a2 fragment of atg3
57 c5oc0A_
not modelled
6.9
12
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome b561;PDBTitle: structure of e. coli superoxide oxidase
58 c4gdkF_
not modelled
6.9
25
PDB header: protein bindingChain: F: PDB Molecule: autophagy-related protein 16-1;PDBTitle: crystal structure of human atg12~atg5 conjugate in complex with an n-2 terminal fragment of atg16l1
59 d1kida_
not modelled
6.9
10
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
60 c6iedA_
not modelled
6.9
9
PDB header: membrane proteinChain: A: PDB Molecule: heme a synthase;PDBTitle: crystal structure of heme a synthase from bacillus subtilis
61 c6e4hA_
not modelled
6.8
33
PDB header: oncoproteinChain: A: PDB Molecule: partner and localizer of brca2;PDBTitle: solution nmr structure of the colied-coil palb2 homodimer
62 c5lnkZ_
not modelled
6.7
19
PDB header: oxidoreductaseChain: Z: PDB Molecule: mitochondrial complex i, pdsw subunit;PDBTitle: entire ovine respiratory complex i
63 d1we3a2
not modelled
6.6
10
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: GroEL apical domain-likeFamily: GroEL-like chaperone, apical domain
64 c5y88R_
not modelled
6.5
23
PDB header: splicingChain: R: PDB Molecule: protein cwc16;PDBTitle: cryo-em structure of the intron-lariat spliceosome ready for2 disassembly from s.cerevisiae at 3.5 angstrom
65 c2jlnA_
not modelled
6.5
5
PDB header: membrane proteinChain: A: PDB Molecule: mhp1;PDBTitle: structure of mhp1, a nucleobase-cation-symport-1 family2 transporter
66 d1z9ha1
not modelled
6.4
13
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain
67 d1hr6a2
not modelled
6.2
6
Fold: LuxS/MPP-like metallohydrolaseSuperfamily: LuxS/MPP-like metallohydrolaseFamily: MPP-like
68 d2gv8a1
not modelled
6.1
12
Fold: FAD/NAD(P)-binding domainSuperfamily: FAD/NAD(P)-binding domainFamily: FAD/NAD-linked reductases, N-terminal and central domains
69 c3j39O_
not modelled
6.0
13
PDB header: ribosomeChain: O: PDB Molecule: 60s ribosomal protein l13a;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
70 c3iz5K_
not modelled
5.9
22
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l13a (l13p);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
71 c3lofF_
not modelled
5.9
16
PDB header: chaperoneChain: F: PDB Molecule: heat shock 70 kda protein 1;PDBTitle: c-terminal domain of human heat shock 70kda protein 1b.
72 c3vviB_
not modelled
5.9
29
PDB header: transport proteinChain: B: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
73 c3vviD_
not modelled
5.9
29
PDB header: transport proteinChain: D: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
74 c3vviE_
not modelled
5.9
29
PDB header: transport proteinChain: E: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
75 c3vviA_
not modelled
5.9
29
PDB header: transport proteinChain: A: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
76 c3vviC_
not modelled
5.9
29
PDB header: transport proteinChain: C: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
77 c3vviH_
not modelled
5.9
29
PDB header: transport proteinChain: H: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
78 c3vviF_
not modelled
5.9
29
PDB header: transport proteinChain: F: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
79 c3vviG_
not modelled
5.9
29
PDB header: transport proteinChain: G: PDB Molecule: non selective cation channel homologous to trp channel;PDBTitle: crystal structure of the coiled-coil domain of the transient receptor2 potential channel from gibberella zeae (trpgz)
80 c2ht2B_
not modelled
5.8
7
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
81 c4btgB_
not modelled
5.8
21
PDB header: virusChain: B: PDB Molecule: major inner protein p1;PDBTitle: coordinates of the bacteriophage phi6 capsid subunits (p1a and p1b)2 fitted into the cryoem reconstruction of the procapsid at 4.4 a3 resolution
82 c5dg3D_
not modelled
5.8
17
PDB header: transferaseChain: D: PDB Molecule: acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-PDBTitle: structure of pseudomonas aeruginosa lpxa in complex with udp-3-o-(r-3-2 hydroxydecanoyl)-glcnac
83 c3vouB_
not modelled
5.7
13
PDB header: transport proteinChain: B: PDB Molecule: ion transport 2 domain protein, voltage-gated sodiumPDBTitle: the crystal structure of nak-navsulp chimera channel
84 d1dxwa_
not modelled
5.6
15
Fold: Trypsin-like serine proteasesSuperfamily: Trypsin-like serine proteasesFamily: Viral proteases
85 c3zf7d_
not modelled
5.6
14
PDB header: ribosomeChain: D: PDB Molecule: PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
86 c2p3xA_
not modelled
5.6
16
PDB header: oxidoreductaseChain: A: PDB Molecule: polyphenol oxidase, chloroplast;PDBTitle: crystal structure of grenache (vitis vinifera) polyphenol2 oxidase
87 d2csba5
not modelled
5.6
38
Fold: Topoisomerase V catalytic domain-likeSuperfamily: Topoisomerase V catalytic domain-likeFamily: Topoisomerase V catalytic domain-like
88 d1b93a_
not modelled
5.5
44
Fold: Methylglyoxal synthase-likeSuperfamily: Methylglyoxal synthase-likeFamily: Methylglyoxal synthase, MgsA
89 c6bl5A_
not modelled
5.5
6
PDB header: viral proteinChain: A: PDB Molecule: head decoration protein;PDBTitle: head decoration protein from the hyperthermophilic phage p74-26
90 c5zfvF_
not modelled
5.5
21
PDB header: transport proteinChain: F: PDB Molecule: 22-mer peptide from biopolymer transport protein exbd;PDBTitle: structure of the exbb/exbd pentameric complex (exbb5exbd1tm)
91 d1l8ya_
not modelled
5.5
18
Fold: HMG-boxSuperfamily: HMG-boxFamily: HMG-box
92 c5f42B_
not modelled
5.5
14
PDB header: transferaseChain: B: PDB Molecule: acyl-[acyl-carrier-protein]--udp-n-acetylglucosamine o-PDBTitle: activity and crystal structure of francisella novicida udp-n-2 acetylglucosamine acyltransferase
93 c5xqgC_
not modelled
5.4
26
PDB header: lyaseChain: C: PDB Molecule: pcrglx protein;PDBTitle: crystal structure of a pl 26 exo-rhamnogalacturonan lyase from2 penicillium chrysogenum complexed with unsaturated galacturonosyl3 rhamnose
94 d1rw2a_
not modelled
5.4
29
Fold: alpha-alpha superhelixSuperfamily: C-terminal domain of Ku80Family: C-terminal domain of Ku80
95 c2f9jP_
not modelled
5.4
30
PDB header: rna binding proteinChain: P: PDB Molecule: splicing factor 3b subunit 1;PDBTitle: 3.0 angstrom resolution structure of a y22m mutant of the spliceosomal2 protein p14 bound to a region of sf3b155
96 d1a0rp_
not modelled
5.3
7
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: Phosducin
97 d1bt3a_
not modelled
5.3
12
Fold: Di-copper centre-containing domainSuperfamily: Di-copper centre-containing domainFamily: Catechol oxidase
98 c4odaD_
not modelled
5.3
13
PDB header: hydrolase/replicationChain: D: PDB Molecule: dna polymerase processivity factor component a20;PDBTitle: crystal structure of the vaccinia virus dna polymerase holoenzyme2 subunit d4 in complex with the a20 n-terminus
99 c3m6zA_
not modelled
5.2
38
PDB header: isomeraseChain: A: PDB Molecule: topoisomerase v;PDBTitle: crystal structure of an n-terminal 44 kda fragment of topoisomerase v2 in the presence of guanidium hydrochloride