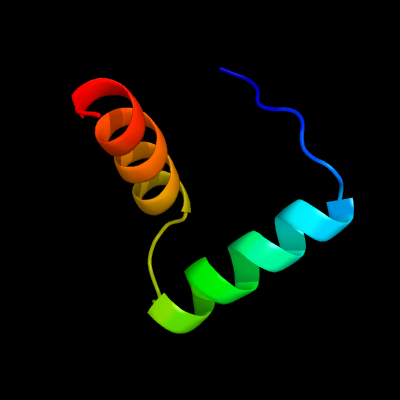
1 c2k5jB_
74.4
21
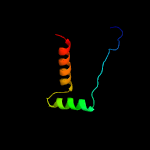
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein yiif;PDBTitle: solution structure of protein yiif from shigella flexneri2 serotype 5b (strain 8401) . northeast structural genomics3 consortium target sft1
2 c2rbfB_
65.2
15
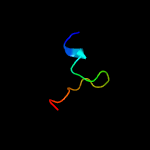
PDB header: oxidoreductase/dnaChain: B: PDB Molecule: bifunctional protein puta;PDBTitle: structure of the ribbon-helix-helix domain of escherichia coli puta2 (puta52) complexed with operator dna (o2)
3 c6iyaD_
63.6
33
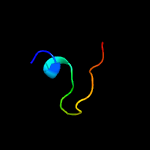
PDB header: antitoxinChain: D: PDB Molecule: transcriptional regulator copg family;PDBTitle: structure of the dna binding domain of antitoxin copaso
4 c2k9iB_
61.8
21
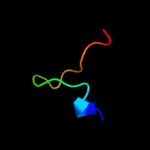
PDB header: dna binding proteinChain: B: PDB Molecule: uncharacterized protein orf56;PDBTitle: nmr structure of plasmid copy control protein orf56 from sulfolobus2 islandicus
5 c5aj3p_
33.9
35
PDB header: ribosomeChain: P: PDB Molecule: mitoribosomal protein bs16m, mrps16;PDBTitle: structure of the small subunit of the mammalian mitoribosome
6 c3e7kF_
31.8
69
PDB header: membrane proteinChain: F: PDB Molecule: trpm7 channel;PDBTitle: crystal structure of an antiparallel coiled-coil tetramerization2 domain from trpm7 channels
7 c3kk4B_
23.1
29
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein bp1543;PDBTitle: uncharacterized protein bp1543 from bordetella pertussis tohama i
8 c3f41B_
22.3
16
PDB header: hydrolaseChain: B: PDB Molecule: phytase;PDBTitle: structure of the tandemly repeated protein tyrosine2 phosphatase like phytase from mitsuokella multacida
9 d2uubr1
21.0
69
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein S18Family: Ribosomal protein S18
10 c5aj3R_
20.9
33
PDB header: ribosomeChain: R: PDB Molecule: mitoribosomal protein bs18m, mrps18c;PDBTitle: structure of the small subunit of the mammalian mitoribosome
11 d2qalr1
20.8
38
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein S18Family: Ribosomal protein S18
12 d1q9ja2
18.6
13
Fold: CoA-dependent acyltransferasesSuperfamily: CoA-dependent acyltransferasesFamily: NRPS condensation domain (amide synthase)
13 c6dzkr_
18.3
41
PDB header: ribosomeChain: R: PDB Molecule: PDBTitle: cryo-em structure of mycobacterium smegmatis c(minus) 30s ribosomal2 subunit with mpy
14 d1i94r_
18.0
65
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Ribosomal protein S18Family: Ribosomal protein S18
15 d1y0na_
17.4
50
Fold: YehU-likeSuperfamily: YehU-likeFamily: YehU-like
16 c5o5jR_
17.1
29
PDB header: ribosomeChain: R: PDB Molecule: 30s ribosomal protein s18 2;PDBTitle: structure of the 30s small ribosomal subunit from mycobacterium2 smegmatis
17 c3j0xU_
16.7
40
PDB header: ribosomeChain: U: PDB Molecule: 30s ribosomal protein s18;PDBTitle: structural characterization of mrna-trna translocation intermediates2 (30s ribosome of class 4b of the six classes)
18 c3df1R_
13.7
38
PDB header: ribosomeChain: R: PDB Molecule: 30s ribosomal protein s18;PDBTitle: crystal structure of the bacterial ribosome from escherichia coli in2 complex with hygromycin b. this file contains the 30s subunit of the3 first 70s ribosome, with hygromycin b bound. the entire crystal4 structure contains two 70s ribosomes.
19 d1h3ob_
13.0
24
Fold: Histone-foldSuperfamily: Histone-foldFamily: TBP-associated factors, TAFs
20 c3bbnR_
12.9
44
PDB header: ribosomeChain: R: PDB Molecule: ribosomal protein s18;PDBTitle: homology model for the spinach chloroplast 30s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome.
21 d2ay0a1
not modelled
12.6
14
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: PutA pre-N-terminal region-like
22 c6hqaK_
not modelled
11.7
40
PDB header: transcriptionChain: K: PDB Molecule: subunit (61/68 kda) of tfiid and saga complexes;PDBTitle: molecular structure of promoter-bound yeast tfiid
23 d2fug21
not modelled
10.6
34
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: NQO2-like
24 c2vsqA_
not modelled
10.2
16
PDB header: ligaseChain: A: PDB Molecule: surfactin synthetase subunit 3;PDBTitle: structure of surfactin a synthetase c (srfa-c), a nonribosomal peptide2 synthetase termination module
25 c5mjyE_
not modelled
9.7
44
PDB header: hydrolaseChain: E: PDB Molecule: zinc finger fyve domain-containing protein 9;PDBTitle: crystal structure of the his domain protein tyrosine phosphatase (hd-2 ptp/ptpn23) bro1 domain (sara complex structure)
26 c5mjyF_
not modelled
9.7
44
PDB header: hydrolaseChain: F: PDB Molecule: zinc finger fyve domain-containing protein 9;PDBTitle: crystal structure of the his domain protein tyrosine phosphatase (hd-2 ptp/ptpn23) bro1 domain (sara complex structure)
27 c6a6xC_
not modelled
9.6
39
PDB header: toxinChain: C: PDB Molecule: antitoxin maze7;PDBTitle: the crystal structure of the mtb maze-mazf-mt9 complex
28 c3wqtB_
not modelled
8.7
24
PDB header: structural genomicsChain: B: PDB Molecule: cell division protein ftsa;PDBTitle: staphylococcus aureus ftsa complexed with amppnp
29 d1u8va2
not modelled
7.5
36
Fold: Acyl-CoA dehydrogenase NM domain-likeSuperfamily: Acyl-CoA dehydrogenase NM domain-likeFamily: Medium chain acyl-CoA dehydrogenase, NM (N-terminal and middle) domains
30 d1tkna_
not modelled
7.1
29
Fold: STAT-likeSuperfamily: CAPPD, an extracellular domain of amyloid beta A4 proteinFamily: CAPPD, an extracellular domain of amyloid beta A4 protein
31 c1qz7B_
not modelled
6.9
75
PDB header: cell adhesionChain: B: PDB Molecule: axin;PDBTitle: beta-catenin binding domain of axin in complex with beta-2 catenin
32 d1wi0a_
not modelled
6.4
37
Fold: beta-Grasp (ubiquitin-like)Superfamily: CAD & PB1 domainsFamily: PB1 domain
33 c4uzzB_
not modelled
6.3
55
PDB header: motor proteinChain: B: PDB Molecule: intraflagellar transporter-like protein;PDBTitle: crystal structure of the ttift52-46 complex
34 c2aj1A_
not modelled
6.3
26
PDB header: hydrolaseChain: A: PDB Molecule: probable cadmium-transporting atpase;PDBTitle: solution structure of apocada
35 c6ckaB_
not modelled
6.2
34
PDB header: viral proteinChain: B: PDB Molecule: paratox;PDBTitle: crystal structure of paratox
36 c4ic7B_
not modelled
6.1
37
PDB header: transferaseChain: B: PDB Molecule: dual specificity mitogen-activated protein kinase kinase 5;PDBTitle: crystal structure of the erk5 kinase domain in complex with an mkk52 binding fragment
37 c6avhA_
not modelled
6.1
14
PDB header: ligase, plant proteinChain: A: PDB Molecule: gh3.15 acyl acid amido synthetase;PDBTitle: gh3.15 acyl acid amido synthetase
38 c1nd9A_
not modelled
5.9
40
PDB header: translationChain: A: PDB Molecule: translation initiation factor if-2;PDBTitle: solution structure of the n-terminal subdomain of2 translation initiation factor if2
39 d1nd9a_
not modelled
5.9
40
Fold: Putative DNA-binding domainSuperfamily: Putative DNA-binding domainFamily: N-terminal subdomain of bacterial translation initiation factor IF2
40 c3k4iC_
not modelled
5.8
15
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein pspto_3204 from2 pseudomonas syringae pv. tomato str. dc3000
41 c4oo2D_
not modelled
5.7
64
PDB header: oxidoreductaseChain: D: PDB Molecule: chlorophenol-4-monooxygenase;PDBTitle: streptomyces globisporus c-1027 fad dependent (s)-3-chloro-β-2 tyrosine-s-sgcc2 c-5 hydroxylase sgcc apo form
42 c1u8vA_
not modelled
5.6
36
PDB header: lyase, isomeraseChain: A: PDB Molecule: gamma-aminobutyrate metabolism dehydratase/isomerase;PDBTitle: crystal structure of 4-hydroxybutyryl-coa dehydratase from clostridium2 aminobutyricum: radical catalysis involving a [4fe-4s] cluster and3 flavin
43 d1jh3a_
not modelled
5.4
44
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Tyrosyl-tRNA synthetase (TyrRS), C-terminal domain
44 c1qeyD_
not modelled
5.3
64
PDB header: gene regulationChain: D: PDB Molecule: protein (regulatory protein mnt);PDBTitle: nmr structure determination of the tetramerization domain2 of the mnt repressor: an asymmetric a-helical assembly in3 slow exchange
45 c1qeyB_
not modelled
5.3
64
PDB header: gene regulationChain: B: PDB Molecule: protein (regulatory protein mnt);PDBTitle: nmr structure determination of the tetramerization domain2 of the mnt repressor: an asymmetric a-helical assembly in3 slow exchange
46 c1qeyA_
not modelled
5.3
64
PDB header: gene regulationChain: A: PDB Molecule: protein (regulatory protein mnt);PDBTitle: nmr structure determination of the tetramerization domain2 of the mnt repressor: an asymmetric a-helical assembly in3 slow exchange
47 c1qeyC_
not modelled
5.3
64
PDB header: gene regulationChain: C: PDB Molecule: protein (regulatory protein mnt);PDBTitle: nmr structure determination of the tetramerization domain2 of the mnt repressor: an asymmetric a-helical assembly in3 slow exchange
48 d2b5id2
not modelled
5.3
36
Fold: Complement control module/SCR domainSuperfamily: Complement control module/SCR domainFamily: Complement control module/SCR domain
49 c2dmeA_
not modelled
5.0
30
PDB header: metal binding proteinChain: A: PDB Molecule: phd finger protein 3;PDBTitle: solution structure of the tfiis domain ii of human phd2 finger protein 3