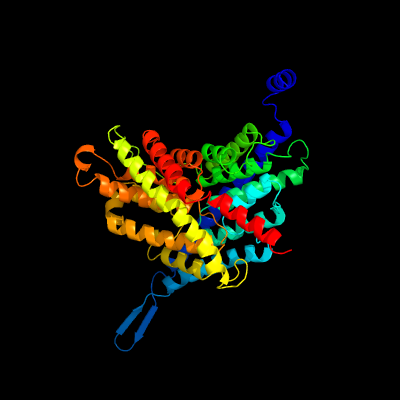
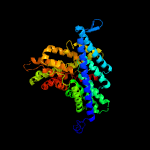
1 d1otsa_
100.0
23
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel
2 c2ht2B_
100.0
23
PDB header: membrane proteinChain: B: PDB Molecule: h(+)/cl(-) exchange transporter clca;PDBTitle: structure of the escherichia coli clc chloride channel2 y445h mutant and fab complex
3 d1kpla_
100.0
25
Fold: Clc chloride channelSuperfamily: Clc chloride channelFamily: Clc chloride channel
4 c3nd0A_
100.0
23
PDB header: transport proteinChain: A: PDB Molecule: sll0855 protein;PDBTitle: x-ray crystal structure of a slow cyanobacterial cl-/h+ antiporter
5 c6coyB_
100.0
19
PDB header: transport proteinChain: B: PDB Molecule: chloride channel protein 1;PDBTitle: human clc-1 chloride ion channel, transmembrane domain
6 c6qvcB_
100.0
18
PDB header: membrane proteinChain: B: PDB Molecule: chloride channel protein 1;PDBTitle: cryoem structure of the human clc-1 chloride channel, cbs state 1
7 c5tr1A_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: chloride channel protein;PDBTitle: cryo-electron microscopy structure of a bovine clc-k chloride channel,2 alternate (class 2) conformation
8 c3orgB_
100.0
22
PDB header: transport proteinChain: B: PDB Molecule: cmclc;PDBTitle: crystal structure of a eukaryotic clc transporter
9 c6d0nA_
100.0
23
PDB header: transport proteinChain: A: PDB Molecule: clc-type fluoride/proton antiporter;PDBTitle: crystal structure of a clc-type fluoride/proton antiporter, v319g2 mutant
10 c3qe7A_
24.4
13
PDB header: transport proteinChain: A: PDB Molecule: uracil permease;PDBTitle: crystal structure of uracil transporter--uraa
11 c2inpE_
15.9
15
PDB header: oxidoreductaseChain: E: PDB Molecule: phenol hydroxylase component pho;PDBTitle: structure of the phenol hydroxylase-regulatory protein2 complex
12 c5xyie_
12.2
10
PDB header: ribosomeChain: E: PDB Molecule: 40s ribosomal protein s4;PDBTitle: small subunit of trichomonas vaginalis ribosome
13 c3m9hB_
10.9
38
PDB header: chaperoneChain: B: PDB Molecule: proteasome-associated atpase;PDBTitle: crystal structure of the amino terminal coiled coil domain of the2 mycobacterium tuberculosis proteasomal atpase mpa
14 c3d9sB_
10.4
18
PDB header: membrane proteinChain: B: PDB Molecule: aquaporin-5;PDBTitle: human aquaporin 5 (aqp5) - high resolution x-ray structure
15 c2kjfA_
9.4
24
PDB header: antimicrobial proteinChain: A: PDB Molecule: carnocyclin-a;PDBTitle: the solution structure of the circular bacteriocin2 carnocyclin a (ccla)
16 c6m97A_
9.2
13
PDB header: transport proteinChain: A: PDB Molecule: chimera protein of high affinity copper uptake protein 1PDBTitle: crystal structure of the high-affinity copper transporter ctr1
17 c2kncA_
9.1
14
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
18 c5cehA_
8.1
40
PDB header: oxidoreductase/inhibitorChain: A: PDB Molecule: lysine-specific demethylase 5a;PDBTitle: structure of histone lysine demethylase kdm5a in complex with2 selective inhibitor
19 d1j4na_
7.8
19
Fold: Aquaporin-likeSuperfamily: Aquaporin-likeFamily: Aquaporin-like
20 c2mgyA_
7.8
11
PDB header: membrane proteinChain: A: PDB Molecule: translocator protein;PDBTitle: solution structure of the mitochondrial translocator protein (tspo) in2 complex with its high-affinity ligand pk11195
21 c3fhkF_
not modelled
7.4
38
PDB header: structural genomics, unknown functionChain: F: PDB Molecule: upf0403 protein yphp;PDBTitle: crystal structure of apc1446, b.subtilis yphp disulfide isomerase
22 c4igpA_
not modelled
7.1
40
PDB header: structural proteinChain: A: PDB Molecule: os05g0196500 protein;PDBTitle: histone h3 lysine 4 demethylating rice jmj703 apo enzyme
23 c3opwA_
not modelled
7.0
30
PDB header: oxidoreductaseChain: A: PDB Molecule: dna damage-responsive transcriptional repressor rph1;PDBTitle: crystal structure of the rph1 catalytic core
24 c5yknA_
not modelled
6.7
40
PDB header: gene regulationChain: A: PDB Molecule: probable lysine-specific demethylase jmj14;PDBTitle: crystal structure of arabidopsis thaliana jmj14 catalytic domain
25 d2qtva4
not modelled
6.7
11
Fold: Gelsolin-likeSuperfamily: C-terminal, gelsolin-like domain of Sec23/24Family: C-terminal, gelsolin-like domain of Sec23/24
26 c5a1fA_
not modelled
6.6
40
PDB header: oxidoreductaseChain: A: PDB Molecule: lysine-specific demethylase 5b, lysine-specificPDBTitle: crystal structure of the catalytic domain of plu1 in complex with2 n-oxalylglycine.
27 c6ip4A_
not modelled
6.5
50
PDB header: gene regulationChain: A: PDB Molecule: arabidopsis jmj13;PDBTitle: crystal structure of arabidopsis thaliana jmj13 catalytic domain in2 complex with nog and an h3k27me3 peptide
28 c5a1sB_
not modelled
6.5
16
PDB header: transport proteinChain: B: PDB Molecule: citrate-sodium symporter;PDBTitle: crystal structure of the sodium-dependent citrate symporter secits2 form salmonella enterica.
29 c4k1cB_
not modelled
6.4
20
PDB header: membrane protein/metal transportChain: B: PDB Molecule: vacuolar calcium ion transporter;PDBTitle: vcx1 calcium/proton exchanger
30 c3twkB_
not modelled
6.4
25
PDB header: hydrolaseChain: B: PDB Molecule: formamidopyrimidine-dna glycosylase 1;PDBTitle: crystal structure of arabidopsis thaliana fpg
31 c2w2iC_
not modelled
6.3
40
PDB header: oxidoreductaseChain: C: PDB Molecule: 2-oxoglutarate oxygenase;PDBTitle: crystal structure of the human 2-oxoglutarate oxygenase2 loc390245
32 c4k1cA_
not modelled
6.1
19
PDB header: membrane protein/metal transportChain: A: PDB Molecule: vacuolar calcium ion transporter;PDBTitle: vcx1 calcium/proton exchanger
33 c5sv9B_
not modelled
5.9
15
PDB header: transport proteinChain: B: PDB Molecule: bor1p boron transporter;PDBTitle: structure of the slc4 transporter bor1p in an inward-facing2 conformation
34 c2os2A_
not modelled
5.5
40
PDB header: oxidoreductaseChain: A: PDB Molecule: jmjc domain-containing histone demethylation protein 3a;PDBTitle: crystal structure of jmjd2a complexed with histone h3 peptide2 trimethylated at lys36
35 c3iz6Z_
not modelled
5.3
14
PDB header: ribosomeChain: Z: PDB Molecule: 40s ribosomal protein s30 (s30e);PDBTitle: localization of the small subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
36 c2q8eB_
not modelled
5.3
40
PDB header: oxidoreductaseChain: B: PDB Molecule: jmjc domain-containing histone demethylation protein 3a;PDBTitle: specificity and mechanism of jmjd2a, a trimethyllysine-specific2 histone demethylase
37 c6hu9l_
not modelled
5.1
25
PDB header: oxidoreductase/electron transportChain: L: PDB Molecule: cytochrome b-c1 complex subunit 1, mitochondrial;PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
38 c6hu9x_
not modelled
5.1
25
PDB header: oxidoreductase/electron transportChain: X: PDB Molecule: PDBTitle: iii2-iv2 mitochondrial respiratory supercomplex from s. cerevisiae
39 c5x1gC_
not modelled
5.0
6
PDB header: structural proteinChain: C: PDB Molecule: wasp homolog-associated protein with actin, membranes andPDBTitle: whamm's microtubule binding motif