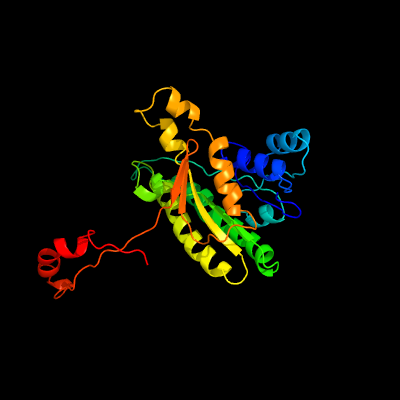
| 1 | c3omlA_
|
|
 |
100.0 |
49 |
PDB header:oxidoreductase, hydrolase
Chain: A: PDB Molecule:peroxisomal multifunctional enzyme type 2, cg3415;
PDBTitle: structure of full-length peroxisomal multifunctional enzyme type 22 from drosophila melanogaster
|
|
|
|
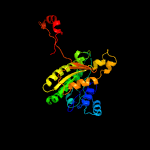
| 2 | c1zbqB_
|
|
 |
100.0 |
49 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:17-beta-hydroxysteroid dehydrogenase 4;
PDBTitle: crystal structure of human 17-beta-hydroxysteroid dehydrogenase type 42 in complex with nad
|
|
|
|
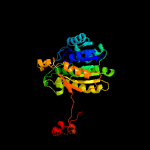
| 3 | d1zbqa1
|
|
 |
100.0 |
49 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
|
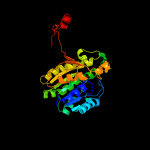
| 4 | c2et6A_
|
|
 |
100.0 |
43 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:(3r)-hydroxyacyl-coa dehydrogenase;
PDBTitle: (3r)-hydroxyacyl-coa dehydrogenase domain of candida tropicalis2 peroxisomal multifunctional enzyme type 2
|
|
|
|
| 5 | d1gz6a_
|
|
 |
100.0 |
49 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
|
| 6 | c3qljB_
|
|
 |
100.0 |
39 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase from mycobacterium2 avium
|
|
|
|
| 7 | c4kzpC_
|
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family
PDBTitle: crystal structure of a putative short chain dehydrogenase from2 mycobacterium smegmatis
|
|
|
|
| 8 | c3t7cC_
|
|
 |
100.0 |
27 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carveol dehydrogenase;
PDBTitle: crystal structure of carveol dehydrogenase from mycobacterium avium2 bound to nad
|
|
|
|
| 9 | c5epoD_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:7-alpha-hydroxysteroid deydrogenase;
PDBTitle: the three-dimensional structure of clostridium absonum 7alpha-2 hydroxysteroid dehydrogenase
|
|
|
|
| 10 | c5jy1C_
|
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of putative short-chain dehydrogenase/reductase from2 burkholderia xenovorans lb400 bound to nad
|
|
|
|
| 11 | c4urfB_
|
|
 |
100.0 |
34 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:cyclohexanol dehydrogenase;
PDBTitle: molecular genetic and crystal structural analysis of 1-(4-2 hydroxyphenyl)-ethanol dehydrogenase from aromatoleum aromaticum ebn1
|
|
|
|
| 12 | c3cxtA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase with different specificities;
PDBTitle: quaternary complex structure of gluconate 5-dehydrogenase from2 streptococcus suis type 2
|
|
|
|
| 13 | c3oecA_
|
|
 |
100.0 |
34 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:carveol dehydrogenase (mytha.01326.c, a0r518 homolog);
PDBTitle: crystal structure of carveol dehydrogenase from mycobacterium2 thermoresistibile
|
|
|
|
| 14 | c4j2hA_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain alcohol dehydrogenase-related dehydrogenase;
PDBTitle: crystal structure of a putative short-chain alcohol dehydrogenase from2 sinorhizobium meliloti 1021 (target nysgrc-011708)
|
|
|
|
| 15 | c3tscB_
|
|
 |
100.0 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of short chain dehydrogenase map_2410 from2 mycobacterium paratuberculosis bound to nad
|
|
|
|
| 16 | d1yxma1
|
|
 |
100.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
|
| 17 | c4lvuB_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: crystal structure of a putative short chain dehydrogenase from2 burkholderia thailandensis
|
|
|
|
| 18 | d2ew8a1
|
|
 |
100.0 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
|
| 19 | c6ci9D_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: rmm microcompartment-associated aminopropanol dehydrogenase nadp +2 aminoacetone holo-structure
|
|
|
|
| 20 | c5o30A_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of the novel halohydrin dehalogenase hheg
|
|
|
|
| 21 | c4afnB_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase fabg;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase2 (fabg) from pseudomonas aeruginosa at 2.3a resolution
|
|
|
| 22 | c5er6C_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: crystal structure of an oxidoreductase from brucella ovis
|
|
|
| 23 | c3uveC_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carveol dehydrogenase ((+)-trans-carveol dehydrogenase);
PDBTitle: crystal structure of carveol dehydrogenase ((+)-trans-carveol2 dehydrogenase) from mycobacterium avium
|
|
|
| 24 | c4nqzF_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh] fabi;
PDBTitle: crystal structure of the pseudomonas aeruginosa enoyl-acyl carrier2 protein reductase (fabi) in apo form
|
|
|
| 25 | c3pgxB_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:carveol dehydrogenase;
PDBTitle: crystal structure of a putative carveol dehydrogenase from2 mycobacterium paratuberculosis bound to nicotinamide adenine3 dinucleotide
|
|
|
| 26 | d1ae1a_ |
|
not modelled |
100.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 27 | c3wtcB_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of gox2036
|
|
|
| 28 | c2p68A_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of aq_1716 from aquifex aeolicus vf5
|
|
|
| 29 | c4nbvA_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase putative short-
PDBTitle: crystal structure of fabg from cupriavidus taiwanensis
|
|
|
| 30 | d1fmca_ |
|
not modelled |
100.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 31 | c4gh5B_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of s-2-hydroxypropyl coenzyme m dehydrogenase (s-2 hpcdh)
|
|
|
| 32 | c4fc6B_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:peroxisomal 2,4-dienoyl-coa reductase;
PDBTitle: studies on dcr shed new light on peroxisomal beta-oxidation: crystal2 structure of the ternary complex of pdcr
|
|
|
| 33 | c3lf2B_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain oxidoreductase q9hya2;
PDBTitle: nadph bound structure of the short chain oxidoreductase q9hya2 from2 pseudomonas aeruginosa pao1 containing an atypical catalytic center
|
|
|
| 34 | d2c07a1 |
|
not modelled |
100.0 |
33 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 35 | c2c07A_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-(acyl-carrier protein) reductase;
PDBTitle: oxoacyl-acp reductase of plasmodium falciparum
|
|
|
| 36 | c5jc8C_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of a putative short-chain dehydrogenase/reductase2 from burkholderia xenovorans
|
|
|
| 37 | c4imrA_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of 3-oxoacyl (acyl-carrier-protein) reductase2 (target efi-506442) from agrobacterium tumefaciens c58 with nadp3 bound
|
|
|
| 38 | d1geea_ |
|
not modelled |
100.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 39 | d2ae2a_ |
|
not modelled |
100.0 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 40 | c3k31B_ |
|
not modelled |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of eonyl-(acyl-carrier-protein) reductase from2 anaplasma phagocytophilum in complex with nad at 1.9a resolution
|
|
|
| 41 | c2jjyD_ |
|
not modelled |
100.0 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of francisella tularensis enoyl reductase2 (ftfabi) with bound nad
|
|
|
| 42 | c3itdA_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:17beta-hydroxysteroid dehydrogenase;
PDBTitle: crystal structure of an inactive 17beta-hydroxysteroid dehydrogenase2 (y167f mutated form) from fungus cochliobolus lunatus
|
|
|
| 43 | c4trrH_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:putative d-beta-hydroxybutyrate dehydrogenase;
PDBTitle: crystal structure of a putative putative d-beta-hydroxybutyrate2 dehydrogenase from burkholderia cenocepacia j2315
|
|
|
| 44 | c2q2qG_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:beta-d-hydroxybutyrate dehydrogenase;
PDBTitle: structure of d-3-hydroxybutyrate dehydrogenase from pseudomonas putida
|
|
|
| 45 | d1zema1 |
|
not modelled |
100.0 |
30 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 46 | c4rgbB_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:carveol dehydrogenase;
PDBTitle: crystal structure of a putative carveol dehydrogenase from2 mycobacterium avium bound to nad
|
|
|
| 47 | c2zatC_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dehydrogenase/reductase sdr family member 4;
PDBTitle: crystal structure of a mammalian reductase
|
|
|
| 48 | c4iboA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:gluconate dehydrogenase;
PDBTitle: crystal structure of a putative gluconate dehydrogenase from2 agrobacterium tumefaciens (target efi-506446)
|
|
|
| 49 | c3afnC_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carbonyl reductase;
PDBTitle: crystal structure of aldose reductase a1-r complexed with nadp
|
|
|
| 50 | c4jroC_ |
|
not modelled |
100.0 |
35 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:fabg protein;
PDBTitle: crystal structure of 3-oxoacyl-[acyl-carrier protein]reductase (fabg)2 from listeria monocytogenes in complex with nadp+
|
|
|
| 51 | c3tzqD_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:short-chain type dehydrogenase/reductase;
PDBTitle: crystal structure of a short-chain type dehydrogenase/reductase from2 mycobacterium marinum
|
|
|
| 52 | c5x8hA_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain dehydrogenase reductase;
PDBTitle: crystal structure of the ketone reductase chkred20 from the genome of2 chryseobacterium sp. ca49
|
|
|
| 53 | d1w6ua_ |
|
not modelled |
100.0 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 54 | d1xhla_ |
|
not modelled |
100.0 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 55 | c3ai3A_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadph-sorbose reductase;
PDBTitle: the crystal structure of l-sorbose reductase from gluconobacter2 frateurii complexed with nadph and l-sorbose
|
|
|
| 56 | c4z9yA_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:2-deoxy-d-gluconate 3-dehydrogenase;
PDBTitle: crystal structure of 2-keto-3-deoxy-d-gluconate dehydrogenase from2 pectobacterium carotovorum
|
|
|
| 57 | c3svtA_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain type dehydrogenase/reductase;
PDBTitle: structure of a short-chain type dehydrogenase/reductase from2 mycobacterium ulcerans
|
|
|
| 58 | c4nimA_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:versicolorin reductase;
PDBTitle: crystal structure of a short chain dehydrogenase from brucella2 melitensis
|
|
|
| 59 | c3grkE_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of short chain dehydrogenase reductase2 sdr glucose-ribitol dehydrogenase from brucella melitensis
|
|
|
| 60 | c4g81A_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative hexonate dehydrogenase;
PDBTitle: crystal structure of a hexonate dehydrogenase ortholog (target efi-2 506402 from salmonella enterica, unliganded structure
|
|
|
| 61 | c3toxG_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: G: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase in complex with2 nad(p) from sinorhizobium meliloti 1021
|
|
|
| 62 | c5u9pB_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gluconate 5-dehydrogenase;
PDBTitle: crystal structure of a gluconate 5-dehydrogenase from burkholderia2 cenocepacia j2315 in complex with nadp and tartrate
|
|
|
| 63 | c3iccA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative 3-oxoacyl-(acyl carrier protein) reductase;
PDBTitle: crystal structure of a putative 3-oxoacyl-(acyl carrier protein)2 reductase from bacillus anthracis at 1.87 a resolution
|
|
|
| 64 | c6d9yB_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of a short chain dehydrogenase/reductase sdr from2 burkholderia phymatum with partially occupied nad
|
|
|
| 65 | c6ds1C_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of cj0485 dehydrogenase in complex with nadp+
|
|
|
| 66 | c4wuvB_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:2-hydroxycyclohexanecarboxyl-coa dehydrogenase;
PDBTitle: crystal structure of a putative d-mannonate oxidoreductase from2 haemophilus influenza (avi_5165, target efi-513796) with bound nad
|
|
|
| 67 | c5h5xH_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:putative oxidoreductase;
PDBTitle: crystal structure of nadh bound carbonyl reductase from streptomyces2 coelicolor
|
|
|
| 68 | c2uvdE_ |
|
not modelled |
100.0 |
38 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: the crystal structure of a 3-oxoacyl-(acyl carrier protein) reductase2 from bacillus anthracis (ba3989)
|
|
|
| 69 | c4m87B_ |
|
not modelled |
100.0 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:enoyl-[acyl-carrier-protein] reductase [nadh];
PDBTitle: crystal structure of enoyl-acyl carrier protein reductase (fabi) from2 neisseria meningitidis in complex with nad+
|
|
|
| 70 | c5k9zB_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of putative short-chain dehydrogenase/reductase from2 burkholderia xenovorans lb400
|
|
|
| 71 | c3r3sD_ |
|
not modelled |
100.0 |
23 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:oxidoreductase;
PDBTitle: structure of the ygha oxidoreductase from salmonella enterica
|
|
|
| 72 | c3o38D_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a short chain dehydrogenase from mycobacterium2 smegmatis
|
|
|
| 73 | d1ja9a_ |
|
not modelled |
100.0 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 74 | c4weoD_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative acetoin(diacetyl) reductase;
PDBTitle: crystal structure of a putative acetoin(diacetyl) reductase2 burkholderia cenocepacia
|
|
|
| 75 | c3rihB_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short chain dehydrogenase or reductase;
PDBTitle: crystal structure of a putative short chain dehydrogenase or reductase2 from mycobacterium abscessus
|
|
|
| 76 | c5ff9C_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:noroxomaritidine/norcraugsodine reductase;
PDBTitle: noroxomaritidine/norcraugsodine reductase in complex with nadp+ and2 tyramine
|
|
|
| 77 | c3gvcB_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable short-chain type dehydrogenase/reductase;
PDBTitle: crystal structure of probable short-chain dehydrogenase-reductase from2 mycobacterium tuberculosis
|
|
|
| 78 | c5itvC_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:dihydroanticapsin 7-dehydrogenase;
PDBTitle: crystal structure of bacillus subtilis bacc dihydroanticapsin 7-2 dehydrogenase in complex with nadh
|
|
|
| 79 | d1hdca_ |
|
not modelled |
100.0 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 80 | c1w4zA_ |
|
not modelled |
100.0 |
31 |
PDB header:antibiotic biosynthesis
Chain: A: PDB Molecule:ketoacyl reductase;
PDBTitle: structure of actinorhodin polyketide (actiii) reductase
|
|
|
| 81 | c5jydA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a putative short chain dehydrogenase from2 burkholderia cenocepacia
|
|
|
| 82 | d1x1ta1 |
|
not modelled |
100.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 83 | c4rziB_ |
|
not modelled |
100.0 |
35 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-ketoacyl-acyl carrier protein reductase;
PDBTitle: crystal structure of phab from synechocystis sp. pcc 6803
|
|
|
| 84 | d1iy8a_ |
|
not modelled |
100.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 85 | c3ijrF_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase family;
PDBTitle: 2.05 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor' in complex3 with nad+
|
|
|
| 86 | c4mowB_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose 1-dehydrogenase;
PDBTitle: crystal structure of a putative glucose 1-dehydrogenase from2 burkholderia cenocepacia j2315
|
|
|
| 87 | c5unlA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-ketoacyl-acp reductase;
PDBTitle: crystal structure of a d-beta-hydroxybutyrate dehydrogenase from2 burkholderia multivorans
|
|
|
| 88 | c3imfA_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase;
PDBTitle: 1.99 angstrom resolution crystal structure of a short chain2 dehydrogenase from bacillus anthracis str. 'ames ancestor'
|
|
|
| 89 | d1nffa_ |
|
not modelled |
100.0 |
29 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 90 | c4egfA_ |
|
not modelled |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-xylulose reductase;
PDBTitle: crystal structure of a l-xylulose reductase from mycobacterium2 smegmatis
|
|
|
| 91 | c6oz7A_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized oxidoreductase yohf;
PDBTitle: putative oxidoreductase from escherichia coli str. k-12
|
|
|
| 92 | c5ojgB_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dehydrogenase/reductase sdr family member 4;
PDBTitle: crystal structure of the dehydrogenase/reductase sdr family member 42 (dhrs4) from caenorhabditis elegans
|
|
|
| 93 | c3lylB_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: structure of 3-oxoacyl-acylcarrier protein reductase, fabg2 from francisella tularensis
|
|
|
| 94 | d1ulua_ |
|
not modelled |
100.0 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 95 | c3uf0A_ |
|
not modelled |
100.0 |
30 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of a putative nad(p) dependent gluconate 5-2 dehydrogenase from beutenbergia cavernae(efi target efi-502044) with3 bound nadp (low occupancy)
|
|
|
| 96 | c3e03C_ |
|
not modelled |
100.0 |
21 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a putative dehydrogenase from xanthomonas2 campestris
|
|
|
| 97 | d2rhca1 |
|
not modelled |
100.0 |
32 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 98 | c2z1nA_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase;
PDBTitle: crystal structure of ape0912 from aeropyrum pernix k1
|
|
|
| 99 | c4gloC_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-oxoacyl-[acyl-carrier protein] reductase;
PDBTitle: crystal structure of a short chain dehydrogenase homolog (target efi-2 505321) from burkholderia multivorans, with bound nad
|
|
|
| 100 | c3sc4A_ |
|
not modelled |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase (a0qtm2 homolog);
PDBTitle: crystal structure of a short chain dehydrogenase (a0qtm2 homolog)2 mycobacterium thermoresistibile
|
|
|
| 101 | c3awdD_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative polyol dehydrogenase;
PDBTitle: crystal structure of gox2181
|
|
|
| 102 | c5tiiC_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:3-oxoacyl-acp reductase;
PDBTitle: comprehensive analysis of a novel ketoreductase for pentangular2 polyphenol biosynthesis
|
|
|
| 103 | c3emkA_ |
|
not modelled |
100.0 |
32 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose/ribitol dehydrogenase;
PDBTitle: 2.5a crystal structure of glucose/ribitol dehydrogenase from brucella2 melitensis
|
|
|
| 104 | c4ni5A_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, short-chain dehydrogenase/reductase family
PDBTitle: crystal structure of a short chain dehydrogenase from brucella suis
|
|
|
| 105 | c4k6fD_ |
|
not modelled |
100.0 |
36 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative acetoacetyl-coa reductase;
PDBTitle: x-ray crystal structure of a putative acetoacetyl-coa reductase from2 burkholderia cenocepacia bound to the co-factor nadp
|
|
|
| 106 | c3ftpD_ |
|
not modelled |
100.0 |
33 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:3-oxoacyl-[acyl-carrier protein] reductase;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase from2 burkholderia pseudomallei at 2.05 a resolution
|
|
|
| 107 | c4dqxB_ |
|
not modelled |
100.0 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable oxidoreductase protein;
PDBTitle: crystal structure of a short chain dehydrogenase from rhizobium etli2 cfn 42
|
|
|
| 108 | d1g0oa_ |
|
not modelled |
100.0 |
27 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 109 | c4hsyA_ |
|
not modelled |
100.0 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ketoreductase siam;
PDBTitle: crystal structure of ketoreductase siam from streptomyces sp. a7248
|
|
|
| 110 | c2yz7B_ |
|
not modelled |
100.0 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:d-3-hydroxybutyrate dehydrogenase;
PDBTitle: x-ray analyses of 3-hydroxybutyrate dehydrogenase from2 alcaligenes faecalis
|
|
|
| 111 | c4fn4A_ |
|
not modelled |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase;
PDBTitle: short-chain nad(h)-dependent dehydrogenase/reductase from sulfolobus2 acidocaldarius
|
|
|
| 112 | c4npcA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sorbitol dehydrogenase;
PDBTitle: crystal structure of an oxidoreductase, short-chain2 dehydrogenase/reductase family protein from brucella suis
|
|
|
| 113 | c3gk3D_ |
|
not modelled |
100.0 |
34 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:acetoacetyl-coa reductase;
PDBTitle: crystal structure of acetoacetyl-coa reductase from burkholderia2 pseudomallei 1710b
|
|
|
| 114 | c4cr8D_ |
|
not modelled |
100.0 |
24 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:n-acylmannosamine 1-dehydrogenase;
PDBTitle: crystal structure of the n-acetyl-d-mannosamine dehydrogenase2 with nad
|
|
|
| 115 | d1xkqa_ |
|
not modelled |
100.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 116 | c3ek2D_ |
|
not modelled |
100.0 |
15 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of eonyl-(acyl carrier protein) reductase from2 burkholderia pseudomallei 1719b
|
|
|
| 117 | c3llsB_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-ketoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of 3-ketoacyl-(acyl-carrier-protein) reductase from2 mycobacterium tuberculosis
|
|
|
| 118 | c4dmmA_ |
|
not modelled |
100.0 |
39 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: 3-oxoacyl-[acyl-carrier-protein] reductase from synechococcus2 elongatus pcc 7942 in complex with nadp
|
|
|
| 119 | c4tkmA_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nadh-dependent reductase for 4-deoxy-l-erythro-5-
PDBTitle: crystal structure of nadh-dependent reductase a1-r' complexed with nad
|
|
|
| 120 | c3v2gA_ |
|
not modelled |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of a dehydrogenase/reductase from sinorhizobium2 meliloti 1021
|
|
|