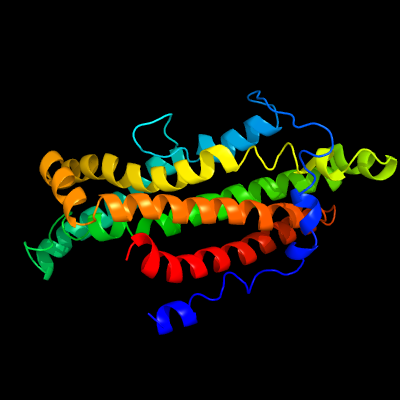
1 c6ic4H_
100.0
21
PDB header: protein transportChain: H: PDB Molecule: abc transporter permease;PDBTitle: cryo-em structure of the a. baumannii mla complex at 8.7 a resolution
2 c5x5yG_
82.7
13
PDB header: membrane proteinChain: G: PDB Molecule: uncharacterized protein;PDBTitle: a membrane protein complex
3 c6mjpG_
65.1
16
PDB header: lipid transportChain: G: PDB Molecule: lps export abc transporter permease lptg;PDBTitle: lptb(e163q)fgc from vibrio cholerae
4 c5l75F_
63.5
12
PDB header: transport proteinChain: F: PDB Molecule: fig000988: predicted permease;PDBTitle: a protein structure
5 c5l75G_
63.1
15
PDB header: transport proteinChain: G: PDB Molecule: fig000906: predicted permease;PDBTitle: a protein structure
6 c5x5yF_
46.8
14
PDB header: membrane proteinChain: F: PDB Molecule: uncharacterized protein;PDBTitle: a membrane protein complex
7 c5xu1M_
16.2
11
PDB header: transport proteinChain: M: PDB Molecule: abc transporter permeae;PDBTitle: structure of a non-canonical abc transporter from streptococcus2 pneumoniae r6
8 c6qvcB_
16.1
11
PDB header: membrane proteinChain: B: PDB Molecule: chloride channel protein 1;PDBTitle: cryoem structure of the human clc-1 chloride channel, cbs state 1
9 d2hafa1
15.7
30
Fold: VC0467-likeSuperfamily: VC0467-likeFamily: VC0467-like
10 c6mjpF_
14.4
13
PDB header: lipid transportChain: F: PDB Molecule: fig000988: predicted permease;PDBTitle: lptb(e163q)fgc from vibrio cholerae
11 d2k0bx1
14.0
27
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain
12 c4migC_
12.3
24
PDB header: oxidoreductaseChain: C: PDB Molecule: pyranose 2-oxidase;PDBTitle: pyranose 2-oxidase from phanerochaete chrysosporium, recombinant wild2 type
13 c2m67A_
12.0
18
PDB header: transport proteinChain: A: PDB Molecule: merf;PDBTitle: full-length mercury transporter protein merf in lipid bilayer2 membranes
14 c2hg5D_
11.7
13
PDB header: membrane proteinChain: D: PDB Molecule: kcsa channel;PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
15 c2hv8D_
11.3
21
PDB header: protein transportChain: D: PDB Molecule: rab11 family-interacting protein 3;PDBTitle: crystal structure of gtp-bound rab11 in complex with fip3
16 c1abzA_
11.0
44
PDB header: de novo designChain: A: PDB Molecule: alpha-t-alpha;PDBTitle: alpha-t-alpha, a de novo designed peptide, nmr, 232 structures
17 c1q7tA_
10.8
36
PDB header: hydrolaseChain: A: PDB Molecule: hypothetical protein rv1170;PDBTitle: rv1170 (mshb) from mycobacterium tuberculosis
18 d1z96a1
10.5
24
Fold: RuvA C-terminal domain-likeSuperfamily: UBA-likeFamily: UBA domain
19 c2aj2A_
10.3
30
PDB header: unknown functionChain: A: PDB Molecule: hypothetical upf0301 protein vc0467;PDBTitle: x-ray crystal structure of protein vc0467 from vibrio2 cholerae. northeast structural genomics consortium target3 vcr8.
20 c2kn8A_
10.0
21
PDB header: protein binding, dna binding proteinChain: A: PDB Molecule: dna cleavage and packaging protein large subunit, ul89;PDBTitle: nmr structure of the c-terminal domain of pul89
21 c5ghaF_
not modelled
10.0
30
PDB header: transferase/transport proteinChain: F: PDB Molecule: sulfur carrier ttub;PDBTitle: sulfur transferase ttua in complex with sulfur carrier ttub
22 c1cf3A_
not modelled
10.0
13
PDB header: oxidoreductase(flavoprotein)Chain: A: PDB Molecule: protein (glucose oxidase);PDBTitle: glucose oxidase from apergillus niger
23 c5sv0C_
not modelled
9.5
18
PDB header: transport proteinChain: C: PDB Molecule: biopolymer transport protein exbb;PDBTitle: structure of the exbb/exbd complex from e. coli at ph 7.0
24 c2jy8A_
not modelled
9.4
27
PDB header: protein bindingChain: A: PDB Molecule: ubiquitin-binding protein p62;PDBTitle: nmr structure of the ubiquitin associated (uba) domain of2 p62 (sqstm1) in complex with ubiquitin. rdc refined
25 c2mfrA_
not modelled
8.6
9
PDB header: transferaseChain: A: PDB Molecule: insulin receptor;PDBTitle: solution structure of the transmembrane domain of the insulin receptor2 in micelles
26 d1q74a_
not modelled
8.1
36
Fold: LmbE-likeSuperfamily: LmbE-likeFamily: LmbE-like
27 c5egiB_
not modelled
7.4
6
PDB header: membrane proteinChain: B: PDB Molecule: uncharacterized protein y57a10a.10;PDBTitle: structure of a trimeric intracellular cation channel from c. elegans2 with bound ca2+
28 c5eikA_
not modelled
6.9
11
PDB header: membrane proteinChain: A: PDB Molecule: uncharacterized protein y57a10a.28;PDBTitle: structure of a trimeric intracellular cation channel from c. elegans2 in the absence of ca2+
29 c3dfmA_
not modelled
6.8
24
PDB header: hydrolaseChain: A: PDB Molecule: teicoplanin pseudoaglycone deacetylase orf2;PDBTitle: the crystal structure of the zinc inhibited form of2 teicoplanin deacetylase orf2
30 c2onkC_
not modelled
6.7
12
PDB header: membrane proteinChain: C: PDB Molecule: molybdate/tungstate abc transporter, permeasePDBTitle: abc transporter modbc in complex with its binding protein2 moda
31 d2onkc1
not modelled
6.7
12
Fold: MetI-likeSuperfamily: MetI-likeFamily: MetI-like
32 c5nikK_
not modelled
6.1
11
PDB header: transport proteinChain: K: PDB Molecule: macrolide export atp-binding/permease protein macb;PDBTitle: structure of the macab-tolc abc-type tripartite multidrug efflux pump
33 c6coyB_
not modelled
6.1
14
PDB header: transport proteinChain: B: PDB Molecule: chloride channel protein 1;PDBTitle: human clc-1 chloride ion channel, transmembrane domain
34 c5l1xH_
not modelled
5.7
32
PDB header: viral proteinChain: H: PDB Molecule: hmpv f1 subunit;PDBTitle: structure of the human metapneumovirus fusion protein in the2 postfusion conformation
35 c6iz0A_
not modelled
5.7
15
PDB header: membrane proteinChain: A: PDB Molecule: trimeric intracellular cation channel type a;PDBTitle: crystal structure analysis of a eukaryotic membrane protein
36 d1i94m_
not modelled
5.3
38
Fold: S13-like H2TH domainSuperfamily: S13-like H2TH domainFamily: Ribosomal protein S13
37 c2m7xA_
not modelled
5.2
14
PDB header: membrane proteinChain: A: PDB Molecule: na(+)/h(+) antiporter;PDBTitle: structural and functional analysis of transmembrane segment iv of the2 salt tolerance protein sod2
38 c4dagA_
not modelled
5.2
32
PDB header: viral protein/immune systemChain: A: PDB Molecule: fusion glycoprotein f0;PDBTitle: structure of the human metapneumovirus fusion protein with2 neutralizing antibody identifies a pneumovirus antigenic site
39 d1g7oa1
not modelled
5.1
16
Fold: GST C-terminal domain-likeSuperfamily: GST C-terminal domain-likeFamily: Glutathione S-transferase (GST), C-terminal domain