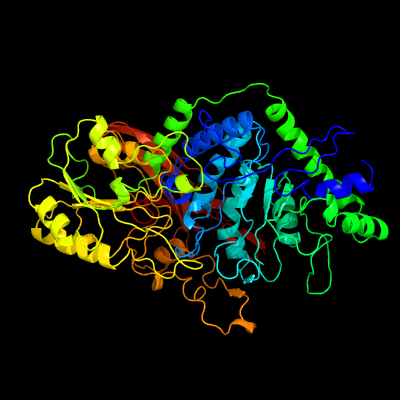
| 1 | c5wabD_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative beta-glucosidase;
PDBTitle: crystal structure of bifidobacterium adolescentis gh3 beta-glucosidase
|
|
|
|
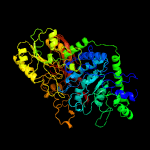
| 2 | c4i3gB_
|
|
 |
100.0 |
39 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of desr, a beta-glucosidase from streptomyces2 venezuelae in complex with d-glucose.
|
|
|
|
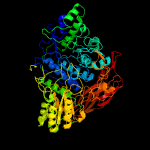
| 3 | c3ac0B_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase i;
PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
|
|
|
|
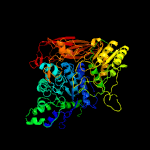
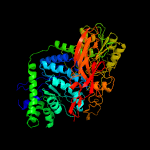
| 4 | c5wabC_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: C: PDB Molecule:putative beta-glucosidase;
PDBTitle: crystal structure of bifidobacterium adolescentis gh3 beta-glucosidase
|
|
|
|
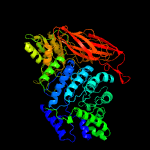
| 5 | c5yotB_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: B: PDB Molecule:isoprimeverose-producing enzyme;
PDBTitle: isoprimeverose-producing enzyme from aspergillus oryzae in complex2 with isoprimeverose
|
|
|
|
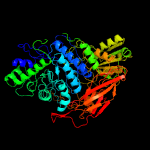
| 6 | c5nbsA_
|
|
 |
100.0 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: structural studies of a glycoside hydrolase family 3 beta-glucosidase2 from the model fungus neurospora crassa
|
|
|
|
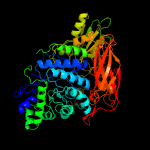
| 7 | c3zz1A_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-d-glucoside glucohydrolase;
PDBTitle: crystal structure of a glycoside hydrolase family 3 beta-glucosidase,2 bgl1 from hypocrea jecorina at 2.1a resolution.
|
|
|
|
| 8 | c4iidB_
|
|
 |
100.0 |
35 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase 1;
PDBTitle: crystal structure of beta-glucosidase 1 from aspergillus aculeatus in2 complex with 1-deoxynojirimycin
|
|
|
|
| 9 | c4d0jD_
|
|
 |
100.0 |
36 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of glycoside hydrolase family 3 beta-2 glucosidase cel3a from the moderately thermophilic fungus3 rasamsonia emersonii
|
|
|
|
| 10 | c2x41A_
|
|
 |
100.0 |
34 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: structure of beta-glucosidase 3b from thermotoga neapolitana in2 complex with glucose
|
|
|
|
| 11 | c5z87B_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:emgh1;
PDBTitle: structural of a novel b-glucosidase emgh1 at 2.3 angstrom from2 erythrobacter marinus
|
|
|
|
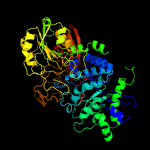
| 12 | c6q7jB_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:exo-1,4-beta-xylosidase xlnd;
PDBTitle: gh3 exo-beta-xylosidase (xlnd) in complex with xylobiose aziridine2 activity based probe
|
|
|
|
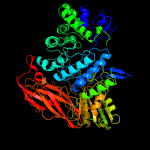
| 13 | c3u48A_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:jmb19063;
PDBTitle: from soil to structure: a novel dimeric family 3-beta-glucosidase2 isolated from compost using metagenomic analysis
|
|
|
|
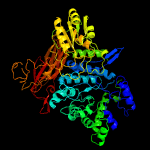
| 14 | c4zo9B_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:lin1840 protein;
PDBTitle: crystal structure of mutant (d270a) beta-glucosidase from listeria2 innocua in complex with laminaribiose
|
|
|
|
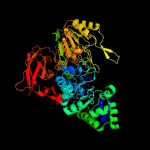
| 15 | c5z9sB_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase family 3 protein;
PDBTitle: functional and structural characterization of a beta-glucosidase2 involved in saponin metabolism from intestinal bacteria
|
|
|
|
| 16 | c5a7mA_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-xylosidase;
PDBTitle: the structure of hypocrea jecorina beta-xylosidase xyl3a (bxl1)
|
|
|
|
| 17 | c5tf0B_
|
|
 |
100.0 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase family 3 n-terminal domain protein;
PDBTitle: crystal structure of glycosil hydrolase family 3 n-terminal domain2 protein from bacteroides intestinalis
|
|
|
|
| 18 | c5jp0A_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase bogh3b;
PDBTitle: bacteroides ovatus xyloglucan pul gh3b with bound glucose
|
|
|
|
| 19 | c3f93D_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of exo-1,3/1,4-beta-glucanase (exop) from2 pseudoalteromonas sp. bb1
|
|
|
|
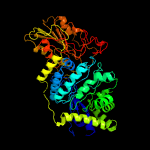
| 20 | c1ex1A_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein (beta-d-glucan exohydrolase isoenzyme exo1);
PDBTitle: beta-d-glucan exohydrolase from barley
|
|
|
|
| 21 | c5m6gA_ |
|
not modelled |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure glucan 1,4-beta-glucosidase from saccharopolyspora2 erythraea
|
|
|
| 22 | c3bmxB_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:uncharacterized lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
|
|
|
| 23 | c3lk6A_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
|
|
|
| 24 | c5vqdA_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucoside phosphorylase bglx;
PDBTitle: beta-glucoside phosphorylase bglx
|
|
|
| 25 | c4zm6A_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase, transferase
Chain: A: PDB Molecule:n-acetyl-beta-d glucosaminidase;
PDBTitle: a unique gcn5-related glucosamine n-acetyltransferase region exist in2 the fungal multi-domain gh3 beta-n-acetylglucosaminidase
|
|
|
| 26 | c3sqlB_ |
|
not modelled |
100.0 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase family 3;
PDBTitle: crystal structure of glycoside hydrolase from synechococcus
|
|
|
| 27 | c5k6lA_ |
|
not modelled |
100.0 |
26 |
PDB header:hydrolase
Chain: A: PDB Molecule:b-glucosidase;
PDBTitle: structure of a gh3 b-glucosidase from cow rumen metagenome
|
|
|
| 28 | c5wvpA_ |
|
not modelled |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: expression, characterization and crystal structure of a novel beta-2 glucosidase from paenibacillus barengoltzii
|
|
|
| 29 | d1x38a1 |
|
not modelled |
100.0 |
28 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:NagZ-like |
|
|
| 30 | c5bzaA_ |
|
not modelled |
100.0 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of cbsa from thermotoga neapolitana
|
|
|
| 31 | c3wo8B_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-n-acetylglucosaminidase;
PDBTitle: crystal structure of the beta-n-acetylglucosaminidase from thermotoga2 maritima
|
|
|
| 32 | c4yyfC_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: the crystal structure of a glycosyl hydrolase of gh3 family member2 from [mycobacterium smegmatis str. mc2 155
|
|
|
| 33 | c5bu9B_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of beta-n-acetylhexosaminidase from beutenbergia2 cavernae dsm 12333
|
|
|
| 34 | c4g6cA_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase 1;
PDBTitle: crystal structure of beta-hexosaminidase 1 from burkholderia2 cenocepacia j2315
|
|
|
| 35 | c5iobC_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-glucosidase-related glycosidases;
PDBTitle: crystal structure of beta-n-acetylglucosaminidase-like protein from2 corynebacterium glutamicum
|
|
|
| 36 | c5g1mA_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of nagz from pseudomonas aeruginosa
|
|
|
| 37 | c3tevA_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyl hyrolase, family 3;
PDBTitle: the crystal structure of glycosyl hydrolase from deinococcus2 radiodurans r1
|
|
|
| 38 | c4gvgB_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of salmonella typhimurium family 3 glycoside2 hydrolase (nagz)
|
|
|
| 39 | d1tr9a_ |
|
not modelled |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:NagZ-like |
|
|
| 40 | d1x38a2 |
|
not modelled |
100.0 |
29 |
Fold:Flavodoxin-like
Superfamily:Beta-D-glucan exohydrolase, C-terminal domain
Family:Beta-D-glucan exohydrolase, C-terminal domain |
|
|
| 41 | d1w8oa1 |
|
not modelled |
97.9 |
22 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:E set domains
Family:E-set domains of sugar-utilizing enzymes |
|
|
| 42 | c2l0dA_ |
|
not modelled |
97.0 |
18 |
PDB header:cell adhesion
Chain: A: PDB Molecule:cell surface protein;
PDBTitle: solution nmr structure of putative cell surface protein ma_4588 (272-2 376 domain) from methanosarcina acetivorans, northeast structural3 genomics consortium target mvr254a
|
|
|
| 43 | c2kl6A_ |
|
not modelled |
96.9 |
20 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of the cardb domain of pf1109 from2 pyrococcus furiosus. northeast structural genomics3 consortium target pfr193a
|
|
|
| 44 | d2q3za2 |
|
not modelled |
96.4 |
13 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
|
|
| 45 | c5vhvB_ |
|
not modelled |
96.2 |
18 |
PDB header:hydrolase/dna
Chain: B: PDB Molecule:alkylpurine dna glycosylase alkc;
PDBTitle: pseudomonas fluorescens alkylpurine dna glycosylase alkc bound to dna2 containing an oxocarbenium-intermediate analog
|
|
|
| 46 | c1l9mB_ |
|
not modelled |
96.0 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:protein-glutamine glutamyltransferase e3;
PDBTitle: three-dimensional structure of the human transglutaminase 32 enzyme: binding of calcium ions change structure for3 activation
|
|
|
| 47 | c4zlgA_ |
|
not modelled |
96.0 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:putative b-glycan phosphorylase;
PDBTitle: cellobionic acid phosphorylase - gluconic acid complex
|
|
|
| 48 | c2x3bB_ |
|
not modelled |
95.6 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:toxic extracellular endopeptidase;
PDBTitle: asap1 inactive mutant e294a, an extracellular toxic zinc2 metalloendopeptidase
|
|
|
| 49 | d1ex0a2 |
|
not modelled |
95.5 |
11 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
|
|
| 50 | d1g0da2 |
|
not modelled |
95.1 |
13 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
|
|
| 51 | d1vjja2 |
|
not modelled |
94.7 |
11 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:Transglutaminase, two C-terminal domains
Family:Transglutaminase, two C-terminal domains |
|
|
| 52 | d2vzsa2 |
|
not modelled |
94.6 |
19 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:beta-Galactosidase/glucuronidase domain
Family:beta-Galactosidase/glucuronidase domain |
|
|
| 53 | c2kutA_ |
|
not modelled |
94.5 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of gmr58a from geobacter metallireducens.2 northeast structural genomics consortium target gmr58a
|
|
|
| 54 | c2qsvA_ |
|
not modelled |
93.4 |
27 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein of unknown function from porphyromonas2 gingivalis w83
|
|
|
| 55 | c4cucA_ |
|
not modelled |
93.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-galactosidase;
PDBTitle: unravelling the multiple functions of the architecturally intricate2 streptococcus pneumoniae beta-galactosidase, bgaa.
|
|
|
| 56 | c3isyA_ |
|
not modelled |
92.4 |
21 |
PDB header:protein binding
Chain: A: PDB Molecule:intracellular proteinase inhibitor;
PDBTitle: crystal structure of an intracellular proteinase inhibitor (ipi,2 bsu11130) from bacillus subtilis at 2.61 a resolution
|
|
|
| 57 | c5z6pB_ |
|
not modelled |
91.9 |
13 |
PDB header:hydrolase
Chain: B: PDB Molecule:b-agarase;
PDBTitle: the crystal structure of an agarase, agwh50c
|
|
|
| 58 | c2h47C_ |
|
not modelled |
91.1 |
18 |
PDB header:oxidoreductase/electron transport
Chain: C: PDB Molecule:azurin;
PDBTitle: crystal structure of an electron transfer complex between2 aromatic amine dephydrogenase and azurin from alcaligenes3 faecalis (form 1)
|
|
|
| 59 | c4bq3A_ |
|
not modelled |
90.9 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:b-agarase;
PDBTitle: structural analysis of an exo-beta-agarase
|
|
|
| 60 | d2ccwa1 |
|
not modelled |
90.2 |
9 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
|
|
| 61 | c3fcsA_ |
|
not modelled |
89.8 |
20 |
PDB header:cell adhesion/blood clotting
Chain: A: PDB Molecule:integrin, alpha 2b;
PDBTitle: structure of complete ectodomain of integrin aiibb3
|
|
|
| 62 | c1kv3F_ |
|
not modelled |
89.0 |
13 |
PDB header:transferase
Chain: F: PDB Molecule:protein-glutamine gamma-glutamyltransferase;
PDBTitle: human tissue transglutaminase in gdp bound form
|
|
|
| 63 | c3rgbA_ |
|
not modelled |
88.3 |
31 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methane monooxygenase subunit b2;
PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
|
|
|
| 64 | d1v7wa2 |
|
not modelled |
88.1 |
30 |
Fold:Supersandwich
Superfamily:Galactose mutarotase-like
Family:Glycosyltransferase family 36 N-terminal domain |
|
|
| 65 | c5t9gD_ |
|
not modelled |
87.7 |
13 |
PDB header:hydrolase
Chain: D: PDB Molecule:glycoside hydrolase;
PDBTitle: crystal structure of bugh2cwt in complex with galactoisofagomine
|
|
|
| 66 | c2cqtA_ |
|
not modelled |
87.1 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:cellobiose phosphorylase;
PDBTitle: crystal structure of cellvibrio gilvus cellobiose phosphorylase2 crystallized from sodium/potassium phosphate
|
|
|
| 67 | c1yewI_ |
|
not modelled |
86.9 |
31 |
PDB header:oxidoreductase, membrane protein
Chain: I: PDB Molecule:particulate methane monooxygenase, b subunit;
PDBTitle: crystal structure of particulate methane monooxygenase
|
|
|
| 68 | c3ay2A_ |
|
not modelled |
86.4 |
12 |
PDB header:antitumor protein, antiviral protein
Chain: A: PDB Molecule:lipid modified azurin protein;
PDBTitle: crystal structure of neisserial azurin
|
|
|
| 69 | d7mdha1 |
|
not modelled |
85.2 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 70 | d1cc3a_ |
|
not modelled |
84.8 |
9 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
|
|
| 71 | c2aanA_ |
|
not modelled |
84.7 |
18 |
PDB header:electron transport
Chain: A: PDB Molecule:auracyanin a;
PDBTitle: auracyanin a: a "blue" copper protein from the green thermophilic2 photosynthetic bacterium,chloroflexus aurantiacus
|
|
|
| 72 | c2a74B_ |
|
not modelled |
84.7 |
14 |
PDB header:immune system
Chain: B: PDB Molecule:complement component c3c;
PDBTitle: human complement component c3c
|
|
|
| 73 | c1v7wA_ |
|
not modelled |
84.1 |
24 |
PDB header:transferase
Chain: A: PDB Molecule:chitobiose phosphorylase;
PDBTitle: crystal structure of vibrio proteolyticus chitobiose phosphorylase in2 complex with glcnac
|
|
|
| 74 | d5mdha1 |
|
not modelled |
84.0 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 75 | d1civa1 |
|
not modelled |
83.6 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 76 | c1g0dA_ |
|
not modelled |
83.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:protein-glutamine gamma-glutamyltransferase;
PDBTitle: crystal structure of red sea bream transglutaminase
|
|
|
| 77 | c2e6jA_ |
|
not modelled |
83.3 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hydin protein;
PDBTitle: solution structure of the c-terminal papd-like domain from2 human hydin protein
|
|
|
| 78 | c2ys4A_ |
|
not modelled |
83.2 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hydrocephalus-inducing protein homolog;
PDBTitle: solution structure of the n-terminal papd-like domain of2 hydin protein from human
|
|
|
| 79 | c1b8vA_ |
|
not modelled |
83.1 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein (malate dehydrogenase);
PDBTitle: malate dehydrogenase from aquaspirillum arcticum
|
|
|
| 80 | c5mdhB_ |
|
not modelled |
83.1 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of ternary complex of porcine cytoplasmic malate2 dehydrogenase alpha-ketomalonate and tnad at 2.4 angstroms resolution
|
|
|
| 81 | c5t98B_ |
|
not modelled |
82.7 |
19 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycoside hydrolase;
PDBTitle: crystal structure of bugh2awt
|
|
|
| 82 | d2co7b1 |
|
not modelled |
82.7 |
29 |
Fold:Immunoglobulin-like beta-sandwich
Superfamily:PapD-like
Family:Pilus chaperone |
|
|
| 83 | d1y7ta1 |
|
not modelled |
82.6 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 84 | d4ubpb_ |
|
not modelled |
82.4 |
38 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
|
|
| 85 | c1f13A_ |
|
not modelled |
82.3 |
13 |
PDB header:coagulation factor
Chain: A: PDB Molecule:cellular coagulation factor xiii zymogen;
PDBTitle: recombinant human cellular coagulation factor xiii
|
|
|
| 86 | d1ejxb_ |
|
not modelled |
82.3 |
40 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
|
|
| 87 | d1b8pa1 |
|
not modelled |
81.6 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 88 | c4acqA_ |
|
not modelled |
80.5 |
15 |
PDB header:hydrolase inhibitor
Chain: A: PDB Molecule:alpha-2-macroglobulin;
PDBTitle: alpha-2 macroglobulin
|
|
|
| 89 | c7mdhA_ |
|
not modelled |
80.4 |
26 |
PDB header:chloroplastic malate dehydrogenase
Chain: A: PDB Molecule:protein (malate dehydrogenase);
PDBTitle: structural basis for light acitvation of a chloroplast enzyme. the2 structure of sorghum nadp-malate dehydrogenase in its oxidized form
|
|
|
| 90 | d1joia_ |
|
not modelled |
79.7 |
13 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
|
|
| 91 | d1nwpa_ |
|
not modelled |
79.3 |
12 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
|
|
| 92 | c4acqC_ |
|
not modelled |
78.9 |
15 |
PDB header:hydrolase inhibitor
Chain: C: PDB Molecule:alpha-2-macroglobulin;
PDBTitle: alpha-2 macroglobulin
|
|
|
| 93 | c4h7pA_ |
|
not modelled |
78.9 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: crystal structure of a putative cytosolic malate dehydrogenase from2 leishmania major friedlin
|
|
|
| 94 | c4fxkB_ |
|
not modelled |
78.4 |
12 |
PDB header:immune system
Chain: B: PDB Molecule:complement c4-a alpha chain;
PDBTitle: human complement c4
|
|
|
| 95 | c5nufA_ |
|
not modelled |
78.1 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase 1, cytoplasmic;
PDBTitle: cytosolic malate dehydrogenase 1
|
|
|
| 96 | c5zi3A_ |
|
not modelled |
77.9 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: mdh3 wild type, apo-form
|
|
|
| 97 | c1wziA_ |
|
not modelled |
77.4 |
28 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: structural basis for alteration of cofactor specificity of malate2 dehydrogenase from thermus flavus
|
|
|
| 98 | c1hyhA_ |
|
not modelled |
77.2 |
20 |
PDB header:oxidoreductase (choh(d)-nad+(a))
Chain: A: PDB Molecule:l-2-hydroxyisocaproate dehydrogenase;
PDBTitle: crystal structure of l-2-hydroxyisocaproate dehydrogenase from2 lactobacillus confusus at 2.2 angstroms resolution-an example of3 strong asymmetry between subunits
|
|
|
| 99 | d1a5za1 |
|
not modelled |
76.8 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 100 | d1llda1 |
|
not modelled |
76.6 |
19 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 101 | d1e9ya1 |
|
not modelled |
76.5 |
38 |
Fold:beta-clip
Superfamily:Urease, beta-subunit
Family:Urease, beta-subunit |
|
|
| 102 | d1azca_ |
|
not modelled |
76.4 |
9 |
Fold:Cupredoxin-like
Superfamily:Cupredoxins
Family:Plastocyanin/azurin-like |
|
|
| 103 | c3zhnA_ |
|
not modelled |
76.3 |
25 |
PDB header:toxin
Chain: A: PDB Molecule:pa_0080;
PDBTitle: crystal structure of the t6ss lipoprotein tssj1 from2 pseudomonas aeruginosa
|
|
|
| 104 | c4uupB_ |
|
not modelled |
75.7 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:malate dehydrogenase;
PDBTitle: reconstructed ancestral trichomonad malate dehydrogenase in2 complex with nadh, so4, and po4
|
|
|
| 105 | c3qgaD_ |
|
not modelled |
74.9 |
30 |
PDB header:hydrolase
Chain: D: PDB Molecule:fusion of urease beta and gamma subunits;
PDBTitle: 3.0 a model of iron containing urease urea2b2 from helicobacter2 mustelae
|
|
|
| 106 | c3qbtH_ |
|
not modelled |
74.6 |
19 |
PDB header:protein transport/hydrolase
Chain: H: PDB Molecule:inositol polyphosphate 5-phosphatase ocrl-1;
PDBTitle: crystal structure of ocrl1 540-678 in complex with rab8a:gppnhp
|
|
|
| 107 | c4uunA_ |
|
not modelled |
74.2 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: trichomonas vaginalis lactate dehydrogenase in complex with nadh
|
|
|
| 108 | c5xyrA_ |
|
not modelled |
74.0 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:chemokine protease c;
PDBTitle: crystal structure of a serine protease from streptococcus species
|
|
|
| 109 | c3rfrI_ |
|
not modelled |
73.9 |
18 |
PDB header:oxidoreductase
Chain: I: PDB Molecule:pmob;
PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
|
|
|
| 110 | d1i0za1 |
|
not modelled |
73.8 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 111 | d2ldxa1 |
|
not modelled |
73.8 |
18 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 112 | d1ldma1 |
|
not modelled |
73.7 |
16 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:LDH N-terminal domain-like |
|
|
| 113 | c3wsvC_ |
|
not modelled |
73.6 |
18 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:l-lactate dehydrogenase;
PDBTitle: crystal structure of minor l-lactate dehydrogenase from enterococcus2 mundtii in the ligands-unbound form
|
|
|
| 114 | c4q3nA_ |
|
not modelled |
73.1 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:mgs-m5;
PDBTitle: crystal structure of mgs-m5, a lactate dehydrogenase enzyme from a2 medee basin deep-sea metagenome library
|
|
|
| 115 | c8ldhA_ |
|
not modelled |
72.8 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:m4 apo-lactate dehydrogenase;
PDBTitle: refined crystal structure of dogfish m4 apo-lactate dehydrogenase
|
|
|
| 116 | c1e1cA_ |
|
not modelled |
72.7 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase alpha chain;
PDBTitle: methylmalonyl-coa mutase h244a mutant
|
|
|
| 117 | c1e9zA_ |
|
not modelled |
71.9 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:urease subunit alpha;
PDBTitle: crystal structure of helicobacter pylori urease
|
|
|
| 118 | c3fn9B_ |
|
not modelled |
71.9 |
8 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative beta-galactosidase;
PDBTitle: crystal structure of putative beta-galactosidase from bacteroides2 fragilis
|
|
|
| 119 | c1mldA_ |
|
not modelled |
71.9 |
20 |
PDB header:oxidoreductase(nad(a)-choh(d))
Chain: A: PDB Molecule:malate dehydrogenase;
PDBTitle: refined structure of mitochondrial malate dehydrogenase2 from porcine heart and the consensus structure for3 dicarboxylic acid oxidoreductases
|
|
|
| 120 | c4djmA_ |
|
not modelled |
71.1 |
21 |
PDB header:chaperone
Chain: A: PDB Molecule:drab;
PDBTitle: crystal structure of the e. coli chaperone drab
|
|
|