| 1 |
|
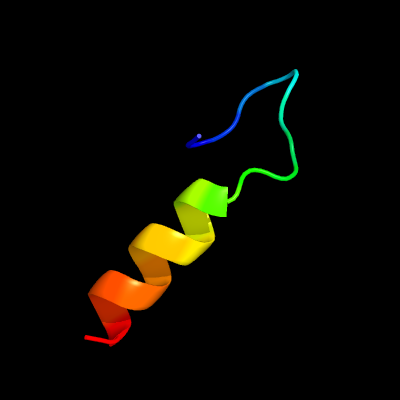
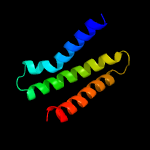
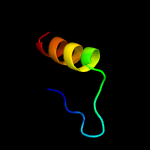
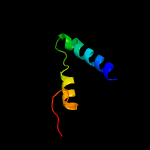
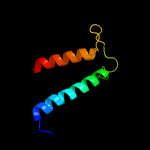
PDB 1qgi chain A
Region: 3 - 24
Aligned: 22
Modelled: 22
Confidence: 26.0%
Identity: 41%
Fold: Lysozyme-like
Superfamily: Lysozyme-like
Family: Chitosanase
Phyre2
| 2 |
|
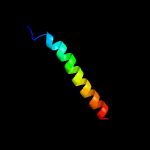
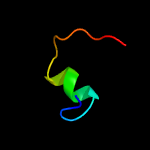
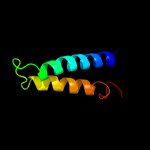
PDB 3klt chain B
Region: 14 - 32
Aligned: 19
Modelled: 19
Confidence: 18.8%
Identity: 16%
PDB header:structural protein
Chain: B: PDB Molecule:vimentin;
PDBTitle: crystal structure of a vimentin fragment
Phyre2
| 3 |
|
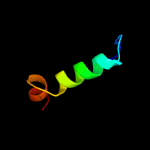
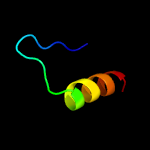
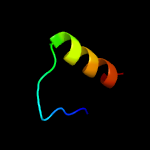
PDB 2yy0 chain D
Region: 14 - 27
Aligned: 14
Modelled: 14
Confidence: 18.0%
Identity: 14%
PDB header:transcription
Chain: D: PDB Molecule:c-myc-binding protein;
PDBTitle: crystal structure of ms0802, c-myc-1 binding protein domain2 from homo sapiens
Phyre2
| 4 |
|
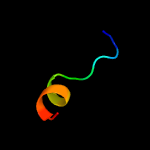
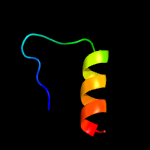
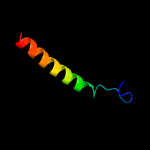
PDB 6iu3 chain A
Region: 29 - 108
Aligned: 79
Modelled: 80
Confidence: 16.5%
Identity: 11%
PDB header:metal transport
Chain: A: PDB Molecule:vit1;
PDBTitle: crystal structure of iron transporter vit1 with zinc ions
Phyre2
| 5 |
|
PDB 2ks1 chain B
Region: 108 - 137
Aligned: 30
Modelled: 30
Confidence: 12.6%
Identity: 20%
PDB header:transferase
Chain: B: PDB Molecule:epidermal growth factor receptor;
PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
Phyre2
| 6 |
|
PDB 2imh chain A domain 1
Region: 8 - 34
Aligned: 27
Modelled: 27
Confidence: 12.5%
Identity: 33%
Fold: Ntn hydrolase-like
Superfamily: N-terminal nucleophile aminohydrolases (Ntn hydrolases)
Family: SPO2555-like
Phyre2
| 7 |
|
PDB 4b1y chain M
Region: 8 - 20
Aligned: 13
Modelled: 13
Confidence: 9.7%
Identity: 31%
PDB header:structural protein
Chain: M: PDB Molecule:phosphatase and actin regulator 1;
PDBTitle: structure of the phactr1 rpel-3 bound to g-actin
Phyre2
| 8 |
|
PDB 2kmu chain A
Region: 4 - 23
Aligned: 20
Modelled: 20
Confidence: 9.3%
Identity: 15%
PDB header:hydrolase
Chain: A: PDB Molecule:atp-dependent dna helicase q4;
PDBTitle: recql4 amino-terminal domain
Phyre2
| 9 |
|
PDB 4ph6 chain A
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 9.2%
Identity: 9%
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: structure of 3-dehydroquinate dehydratase from enterococcus faecalis
Phyre2
| 10 |
|
PDB 2imr chain A domain 1
Region: 9 - 28
Aligned: 18
Modelled: 20
Confidence: 9.1%
Identity: 44%
Fold: Composite domain of metallo-dependent hydrolases
Superfamily: Composite domain of metallo-dependent hydrolases
Family: DR0824-like
Phyre2
| 11 |
|
PDB 5kbh chain B
Region: 10 - 20
Aligned: 11
Modelled: 11
Confidence: 8.4%
Identity: 27%
PDB header:transcription
Chain: B: PDB Molecule:mopr;
PDBTitle: crystal structure of the aromatic sensor domain of mopr in complex2 with 3-chloro-phenol
Phyre2
| 12 |
|
PDB 3js3 chain C
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 8.3%
Identity: 13%
PDB header:lyase
Chain: C: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of type i 3-dehydroquinate dehydratase (arod) from2 clostridium difficile with covalent reaction intermediate
Phyre2
| 13 |
|
PDB 2yr1 chain B
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 7.4%
Identity: 22%
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: crystal structure of 3-dehydroquinate dehydratase from geobacillus2 kaustophilus hta426
Phyre2
| 14 |
|
PDB 6b4g chain G
Region: 66 - 111
Aligned: 45
Modelled: 46
Confidence: 7.2%
Identity: 16%
PDB header:transport protein
Chain: G: PDB Molecule:nucleoporin gle1;
PDBTitle: crystal structure of chaetomium thermophilum gle1 ctd-nup42 gbm2 complex
Phyre2
| 15 |
|
PDB 3l2i chain B
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 7.2%
Identity: 9%
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate dehydratase;
PDBTitle: 1.85 angstrom crystal structure of the 3-dehydroquinate dehydratase2 (arod) from salmonella typhimurium lt2.
Phyre2
| 16 |
|
PDB 2mmu chain A
Region: 32 - 85
Aligned: 48
Modelled: 54
Confidence: 7.1%
Identity: 19%
PDB header:cell cycle
Chain: A: PDB Molecule:cell division protein crga;
PDBTitle: structure of crga, a cell division structural and regulatory protein2 from mycobacterium tuberculosis, in lipid bilayers
Phyre2
| 17 |
|
PDB 3odm chain E
Region: 1 - 13
Aligned: 13
Modelled: 13
Confidence: 7.1%
Identity: 46%
PDB header:lyase
Chain: E: PDB Molecule:phosphoenolpyruvate carboxylase;
PDBTitle: archaeal-type phosphoenolpyruvate carboxylase
Phyre2
| 18 |
|
PDB 1sfl chain A
Region: 4 - 26
Aligned: 23
Modelled: 23
Confidence: 6.9%
Identity: 22%
Fold: TIM beta/alpha-barrel
Superfamily: Aldolase
Family: Class I aldolase
Phyre2
| 19 |
|
PDB 3sjr chain B
Region: 9 - 42
Aligned: 34
Modelled: 34
Confidence: 6.6%
Identity: 29%
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of conserved unkown function protein cv_1783 from2 chromobacterium violaceum atcc 12472
Phyre2
| 20 |
|
PDB 2vv5 chain A domain 3
Region: 18 - 80
Aligned: 55
Modelled: 63
Confidence: 6.5%
Identity: 20%
Fold: Mechanosensitive channel protein MscS (YggB), transmembrane region
Superfamily: Mechanosensitive channel protein MscS (YggB), transmembrane region
Family: Mechanosensitive channel protein MscS (YggB), transmembrane region
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|