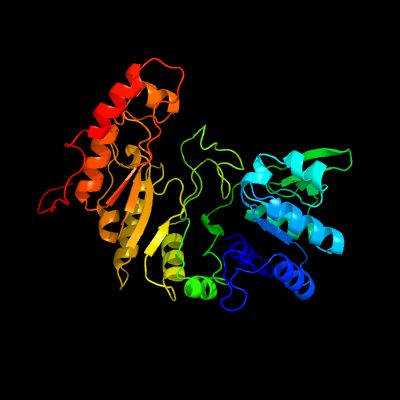
| 1 | c5ul4A_
|
|
 |
100.0 |
25 |
PDB header:metal binding protein
Chain: A: PDB Molecule:oxsb protein;
PDBTitle: structure of cobalamin-dependent s-adenosylmethionine radical enzyme2 oxsb with aqua-cobalamin and s-adenosylmethionine bound
|
|
|
|
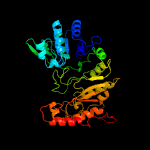
| 2 | c6fd2B_
|
|
 |
100.0 |
18 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:putative apramycin biosynthetic oxidoreductase 4;
PDBTitle: radical sam 1,2-diol dehydratase aprd4 in complex with its substrate2 paromamine
|
|
|
|
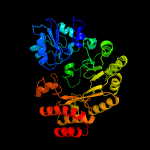
| 3 | c4jc0B_
|
|
 |
100.0 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:ribosomal protein s12 methylthiotransferase rimo;
PDBTitle: crystal structure of thermotoga maritima holo rimo in complex with2 pentasulfide, northeast structural genomics consortium target vr77
|
|
|
|
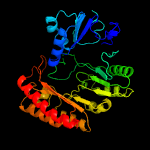
| 4 | c5l7jA_
|
|
 |
100.0 |
19 |
PDB header:translation
Chain: A: PDB Molecule:elp3 family;
PDBTitle: crystal structure of elp3 from dehalococcoides mccartyi
|
|
|
|
| 5 | c2qgqF_
|
|
 |
100.0 |
9 |
PDB header:structural genomics, unknown function
Chain: F: PDB Molecule:protein tm_1862;
PDBTitle: crystal structure of tm_1862 from thermotoga maritima. northeast2 structural genomics consortium target vr77
|
|
|
|
| 6 | c3cixA_
|
|
 |
100.0 |
18 |
PDB header:adomet binding protein
Chain: A: PDB Molecule:fefe-hydrogenase maturase;
PDBTitle: x-ray structure of the [fefe]-hydrogenase maturase hyde from2 thermotoga maritima in complex with thiocyanate
|
|
|
|
| 7 | d1olta_
|
|
 |
100.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Oxygen-independent coproporphyrinogen III oxidase HemN |
|
|
|
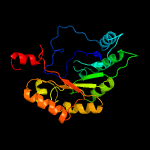
| 8 | c6qk7C_
|
|
 |
99.9 |
18 |
PDB header:translation
Chain: C: PDB Molecule:elongator complex protein 3;
PDBTitle: elongator catalytic subcomplex elp123 lobe
|
|
|
|
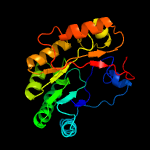
| 9 | d1r30a_
|
|
 |
99.9 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
|
|
|
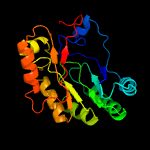
| 10 | c1r30A_
|
|
 |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-adenosylmethionine-2 dependent radical enzyme
|
|
|
|
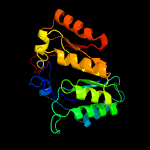
| 11 | c3t7vA_
|
|
 |
99.9 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:methylornithine synthase pylb;
PDBTitle: crystal structure of methylornithine synthase (pylb)
|
|
|
|
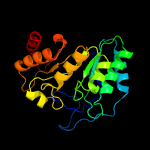
| 12 | c5exkG_
|
|
 |
99.9 |
15 |
PDB header:transferase
Chain: G: PDB Molecule:lipoyl synthase;
PDBTitle: crystal structure of m. tuberculosis lipoyl synthase with 6-2 thiooctanoyl peptide intermediate
|
|
|
|
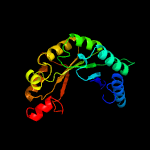
| 13 | c6iazA_
|
|
 |
99.9 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:histone acetyltransferase, elp3 family;
PDBTitle: the archaeal methanocaldococcus infernus elp3 with n-terminus deletion2 (1-46)
|
|
|
|
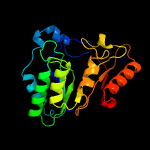
| 14 | c4u0pB_
|
|
 |
99.8 |
13 |
PDB header:transferase
Chain: B: PDB Molecule:lipoyl synthase 2;
PDBTitle: the crystal structure of lipoyl synthase in complex with s-adenosyl2 homocysteine
|
|
|
|
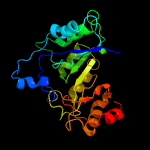
| 15 | c3rfaA_
|
|
 |
99.8 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ribosomal rna large subunit methyltransferase n;
PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
|
|
|
|
| 16 | c4wcxC_
|
|
 |
99.8 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:biotin and thiamin synthesis associated;
PDBTitle: crystal structure of hydg: a maturase of the [fefe]-hydrogenase
|
|
|
|
| 17 | c4rtbA_
|
|
 |
99.7 |
16 |
PDB header:lyase
Chain: A: PDB Molecule:hydg protein;
PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
|
|
|
|
| 18 | c3rfaB_
|
|
 |
99.7 |
14 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ribosomal rna large subunit methyltransferase n;
PDBTitle: x-ray structure of rlmn from escherichia coli in complex with s-2 adenosylmethionine
|
|
|
|
| 19 | c4fheA_
|
|
 |
99.6 |
7 |
PDB header:lyase
Chain: A: PDB Molecule:spore photoproduct lyase;
PDBTitle: spore photoproduct lyase c140a mutant
|
|
|
|
| 20 | c6fz6B_
|
|
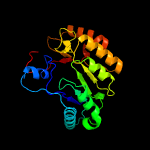 |
99.6 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:probable dual-specificity rna methyltransferase rlmn;
PDBTitle: crystal structure of a radical sam methyltransferase from2 sphaerobacter thermophilus
|
|
|
|
| 21 | c4r33A_ |
|
not modelled |
99.6 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:nosl;
PDBTitle: x-ray structure of the tryptophan lyase nosl with tryptophan and s-2 adenosyl-l-homocysteine bound
|
|
|
| 22 | d1tv8a_ |
|
not modelled |
99.5 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:MoCo biosynthesis proteins |
|
|
| 23 | c5vslB_ |
|
not modelled |
99.5 |
8 |
PDB header:antiviral protein
Chain: B: PDB Molecule:radical s-adenosyl methionine domain-containing protein 2;
PDBTitle: crystal structure of viperin with bound [4fe-4s] cluster and s-2 adenosylhomocysteine (sah)
|
|
|
| 24 | c5v1tA_ |
|
not modelled |
99.5 |
10 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam;
PDBTitle: crystal structure of streptococcus suis suib bound to precursor2 peptide suia
|
|
|
| 25 | c2yx0A_ |
|
not modelled |
99.5 |
10 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam enzyme;
PDBTitle: crystal structure of p. horikoshii tyw1
|
|
|
| 26 | c3c8fA_ |
|
not modelled |
99.5 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyruvate formate-lyase 1-activating enzyme;
PDBTitle: 4fe-4s-pyruvate formate-lyase activating enzyme with partially2 disordered adomet
|
|
|
| 27 | c2a5hC_ |
|
not modelled |
99.3 |
13 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
|
|
| 28 | c6b4cH_ |
|
not modelled |
99.2 |
8 |
PDB header:antiviral protein
Chain: H: PDB Molecule:viperin;
PDBTitle: structure of viperin from trichoderma virens
|
|
|
| 29 | c6efnA_ |
|
not modelled |
99.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sporulation killing factor maturation protein skfb;
PDBTitle: structure of a ripp maturase, skfb
|
|
|
| 30 | c4k39A_ |
|
not modelled |
99.1 |
8 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:anaerobic sulfatase-maturating enzyme;
PDBTitle: native ansmecpe with bound adomet and cp18cys peptide
|
|
|
| 31 | c5th5C_ |
|
not modelled |
99.0 |
12 |
PDB header:lyase
Chain: C: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from bacillus subtilis with 6-carboxypterin-2 5'-deoxyadenosyl ester bound
|
|
|
| 32 | c4m7tA_ |
|
not modelled |
99.0 |
11 |
PDB header:metal binding protein
Chain: A: PDB Molecule:btrn;
PDBTitle: crystal structure of btrn in complex with adomet and 2-doia
|
|
|
| 33 | d3bula2 |
|
not modelled |
98.6 |
16 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 34 | c5wggA_ |
|
not modelled |
98.5 |
8 |
PDB header:peptide binding protein
Chain: A: PDB Molecule:radical sam domain protein;
PDBTitle: structural insights into thioether bond formation in the biosynthesis2 of sactipeptides
|
|
|
| 35 | c1bmtB_ |
|
not modelled |
98.4 |
16 |
PDB header:methyltransferase
Chain: B: PDB Molecule:methionine synthase;
PDBTitle: how a protein binds b12: a 3.o angstrom x-ray structure of2 the b12-binding domains of methionine synthase
|
|
|
| 36 | c1y80A_ |
|
not modelled |
98.3 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:predicted cobalamin binding protein;
PDBTitle: structure of a corrinoid (factor iiim)-binding protein from moorella2 thermoacetica
|
|
|
| 37 | c4jgiB_ |
|
not modelled |
98.0 |
14 |
PDB header:protein binding
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: 1.5 angstrom crystal structure of a novel cobalamin-binding protein2 from desulfitobacterium hafniense dcb-2
|
|
|
| 38 | c6nhlB_ |
|
not modelled |
97.8 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from escherichia coli
|
|
|
| 39 | c4hh3C_ |
|
not modelled |
97.8 |
13 |
PDB header:flavoprotein/transcription
Chain: C: PDB Molecule:appa protein;
PDBTitle: structure of the appa-ppsr2 core complex from rb. sphaeroides
|
|
|
| 40 | c2i2xD_ |
|
not modelled |
97.8 |
18 |
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
|
|
|
| 41 | d1ccwa_ |
|
not modelled |
97.8 |
16 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 42 | d7reqa2 |
|
not modelled |
97.7 |
14 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 43 | d1xrsb1 |
|
not modelled |
97.7 |
14 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 44 | c1k98A_ |
|
not modelled |
97.7 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:methionine synthase;
PDBTitle: adomet complex of meth c-terminal fragment
|
|
|
| 45 | c2z2uA_ |
|
not modelled |
97.6 |
10 |
PDB header:metal binding protein
Chain: A: PDB Molecule:upf0026 protein mj0257;
PDBTitle: crystal structure of archaeal tyw1
|
|
|
| 46 | c2yxbA_ |
|
not modelled |
97.6 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
|
|
| 47 | c3ezxA_ |
|
not modelled |
97.5 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:monomethylamine corrinoid protein 1;
PDBTitle: structure of methanosarcina barkeri monomethylamine2 corrinoid protein
|
|
|
| 48 | c4njkA_ |
|
not modelled |
97.5 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from burkholderia multivorans in complex2 with adomet, 7-carboxy-7-deazaguanine, and mg2+
|
|
|
| 49 | c4r3uD_ |
|
not modelled |
97.5 |
13 |
PDB header:isomerase
Chain: D: PDB Molecule:2-hydroxyisobutyryl-coa mutase small subunit;
PDBTitle: crystal structure of 2-hydroxyisobutyryl-coa mutase
|
|
|
| 50 | c1xrsB_ |
|
not modelled |
97.3 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:d-lysine 5,6-aminomutase beta subunit;
PDBTitle: crystal structure of lysine 5,6-aminomutase in complex with plp,2 cobalamin, and 5'-deoxyadenosine
|
|
|
| 51 | c3canA_ |
|
not modelled |
97.2 |
8 |
PDB header:lyase activator
Chain: A: PDB Molecule:pyruvate-formate lyase-activating enzyme;
PDBTitle: crystal structure of a domain of pyruvate-formate lyase-activating2 enzyme from bacteroides vulgatus atcc 8482
|
|
|
| 52 | d1fmfa_ |
|
not modelled |
97.2 |
13 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 53 | c4hh0B_ |
|
not modelled |
97.1 |
15 |
PDB header:flavoprotein,signaling protein
Chain: B: PDB Molecule:appa protein;
PDBTitle: dark-state structure of appa c20s without the cys-rich region from rb.2 sphaeroides
|
|
|
| 54 | c3whpA_ |
|
not modelled |
96.3 |
19 |
PDB header:gene regulation
Chain: A: PDB Molecule:probable transcriptional regulator;
PDBTitle: crystal structure of the c-terminal domain of themus thermophilus litr2 in complex with cobalamin
|
|
|
| 55 | c1e1cA_ |
|
not modelled |
95.6 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase alpha chain;
PDBTitle: methylmalonyl-coa mutase h244a mutant
|
|
|
| 56 | c6ifhA_ |
|
not modelled |
95.4 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:sporulation initiation phosphotransferase f;
PDBTitle: unphosphorylated spo0f from paenisporosarcina sp. tg-14
|
|
|
| 57 | d1mvoa_ |
|
not modelled |
95.2 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 58 | d1kgsa2 |
|
not modelled |
94.8 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 59 | c1ys7B_ |
|
not modelled |
94.7 |
13 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulatory protein prra;
PDBTitle: crystal structure of the response regulator protein prra complexed2 with mg2+
|
|
|
| 60 | c3dzdA_ |
|
not modelled |
94.7 |
6 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: crystal structure of sigma54 activator ntrc4 in the inactive2 state
|
|
|
| 61 | c3rmjB_ |
|
not modelled |
94.6 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:2-isopropylmalate synthase;
PDBTitle: crystal structure of truncated alpha-isopropylmalate synthase from2 neisseria meningitidis
|
|
|
| 62 | d1ny5a1 |
|
not modelled |
94.6 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 63 | d1ys7a2 |
|
not modelled |
94.5 |
14 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 64 | c2jrlA_ |
|
not modelled |
94.4 |
6 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: solution structure of the beryllofluoride-activated ntrc4 receiver2 domain dimer
|
|
|
| 65 | c6c8vA_ |
|
not modelled |
94.4 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme pqq synthesis protein e;
PDBTitle: x-ray structure of pqqe from methylobacterium extorquens
|
|
|
| 66 | c3bleA_ |
|
not modelled |
94.4 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:citramalate synthase from leptospira interrogans;
PDBTitle: crystal structure of the catalytic domain of licms in complexed with2 malonate
|
|
|
| 67 | c2rjnA_ |
|
not modelled |
94.2 |
11 |
PDB header:hydrolase
Chain: A: PDB Molecule:response regulator receiver:metal-dependent
PDBTitle: crystal structure of an uncharacterized protein q2bku2 from2 neptuniibacter caesariensis
|
|
|
| 68 | c5uicA_ |
|
not modelled |
94.0 |
16 |
PDB header:transcription
Chain: A: PDB Molecule:two-component response regulator;
PDBTitle: structure of the francisella response regulator receiver domain, qseb
|
|
|
| 69 | c5ep0A_ |
|
not modelled |
93.8 |
15 |
PDB header:transcription
Chain: A: PDB Molecule:putative repressor protein luxo;
PDBTitle: quorum-sensing signal integrator luxo - receiver+catalytic domains
|
|
|
| 70 | c3cfyA_ |
|
not modelled |
93.7 |
12 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:putative luxo repressor protein;
PDBTitle: crystal structure of signal receiver domain of putative luxo repressor2 protein from vibrio parahaemolyticus
|
|
|
| 71 | c3hdgE_ |
|
not modelled |
93.6 |
8 |
PDB header:structural genomics, unknown function
Chain: E: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of the n-terminal domain of an2 uncharacterized protein (ws1339) from wolinella3 succinogenes
|
|
|
| 72 | d2pl1a1 |
|
not modelled |
93.5 |
16 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 73 | c4q7eA_ |
|
not modelled |
93.4 |
10 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator of a two component regulatory system;
PDBTitle: non-phosphorylated hemr receiver domain from leptospira biflexa
|
|
|
| 74 | d1yioa2 |
|
not modelled |
93.4 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 75 | d1xhfa1 |
|
not modelled |
93.3 |
16 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 76 | d1peya_ |
|
not modelled |
93.3 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 77 | c1ny5A_ |
|
not modelled |
93.2 |
10 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulator (ntrc family);
PDBTitle: crystal structure of sigm54 activator (aaa+ atpase) in the inactive2 state
|
|
|
| 78 | d2a9pa1 |
|
not modelled |
93.2 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 79 | c4qpjC_ |
|
not modelled |
93.1 |
13 |
PDB header:signaling protein/dna binding protein
Chain: C: PDB Molecule:cell cycle response regulator ctra;
PDBTitle: 2.7 angstrom structure of a phosphotransferase in complex with a2 receiver domain
|
|
|
| 80 | c3f6cB_ |
|
not modelled |
93.0 |
13 |
PDB header:dna binding protein
Chain: B: PDB Molecule:positive transcription regulator evga;
PDBTitle: crystal structure of n-terminal domain of positive transcription2 regulator evga from escherichia coli
|
|
|
| 81 | c3w9sB_ |
|
not modelled |
93.0 |
16 |
PDB header:signaling protein/antimicrobial protein
Chain: B: PDB Molecule:ompr family response regulator in two-component regulatory
PDBTitle: crystal structure analysis of the n-terminal receiver domain of2 response regulator pmra
|
|
|
| 82 | c2zwmA_ |
|
not modelled |
93.0 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:transcriptional regulatory protein yycf;
PDBTitle: crystal structure of yycf receiver domain from bacillus2 subtilis
|
|
|
| 83 | d1dbwa_ |
|
not modelled |
93.0 |
10 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 84 | d1krwa_ |
|
not modelled |
92.7 |
19 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 85 | c2ayxA_ |
|
not modelled |
92.5 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:sensor kinase protein rcsc;
PDBTitle: solution structure of the e.coli rcsc c-terminus (residues2 700-949) containing linker region and phosphoreceiver3 domain
|
|
|
| 86 | d2ayxa1 |
|
not modelled |
92.5 |
12 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 87 | c2ftpA_ |
|
not modelled |
92.5 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:hydroxymethylglutaryl-coa lyase;
PDBTitle: crystal structure of hydroxymethylglutaryl-coa lyase from pseudomonas2 aeruginosa
|
|
|
| 88 | c5c8eC_ |
|
not modelled |
92.5 |
16 |
PDB header:transcription regulator/dna
Chain: C: PDB Molecule:light-dependent transcriptional regulator carh;
PDBTitle: crystal structure of thermus thermophilus carh bound to2 adenosylcobalamin and a 26-bp dna segment
|
|
|
| 89 | c5u8mA_ |
|
not modelled |
92.1 |
13 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator;
PDBTitle: a novel family of redox sensors in the streptococci evolved from two-2 component response regulators
|
|
|
| 90 | c3nhzA_ |
|
not modelled |
92.0 |
16 |
PDB header:dna binding protein
Chain: A: PDB Molecule:two component system transcriptional regulator mtra;
PDBTitle: structure of n-terminal domain of mtra
|
|
|
| 91 | c3bicA_ |
|
not modelled |
91.9 |
15 |
PDB header:isomerase
Chain: A: PDB Molecule:methylmalonyl-coa mutase, mitochondrial precursor;
PDBTitle: crystal structure of human methylmalonyl-coa mutase
|
|
|
| 92 | c4lzlA_ |
|
not modelled |
91.8 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator;
PDBTitle: structure of the inactive form of the regulatory domain from the2 repressor of iron transport regulator (ritr)
|
|
|
| 93 | d1qkka_ |
|
not modelled |
91.5 |
11 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 94 | d1zesa1 |
|
not modelled |
91.5 |
9 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 95 | c3crnA_ |
|
not modelled |
91.2 |
14 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver domain protein, chey-like;
PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
|
|
|
| 96 | c1zn2A_ |
|
not modelled |
91.2 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:response regulatory protein;
PDBTitle: low resolution structure of response regulator styr
|
|
|
| 97 | c3rqiA_ |
|
not modelled |
91.0 |
12 |
PDB header:transcription
Chain: A: PDB Molecule:response regulator protein;
PDBTitle: crystal structure of a response regulator protein from burkholderia2 pseudomallei with a phosphorylated aspartic acid, calcium ion and3 citrate
|
|
|
| 98 | c3hv2B_ |
|
not modelled |
90.9 |
8 |
PDB header:signaling protein
Chain: B: PDB Molecule:response regulator/hd domain protein;
PDBTitle: crystal structure of signal receiver domain of hd domain-containing2 protein from pseudomonas fluorescens pf-5
|
|
|
| 99 | c2oqrA_ |
|
not modelled |
90.9 |
13 |
PDB header:transcription,signaling protein
Chain: A: PDB Molecule:sensory transduction protein regx3;
PDBTitle: the structure of the response regulator regx3 from mycobacterium2 tuberculosis
|
|
|
| 100 | d1h4pa_ |
|
not modelled |
90.7 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
|
|
| 101 | c2qr3A_ |
|
not modelled |
90.7 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:two-component system response regulator;
PDBTitle: crystal structure of the n-terminal signal receiver domain of two-2 component system response regulator from bacteroides fragilis
|
|
|
| 102 | c4s05B_ |
|
not modelled |
90.7 |
17 |
PDB header:transcription/dna
Chain: B: PDB Molecule:dna-binding transcriptional regulator basr;
PDBTitle: crystal structure of klebsiella pneumoniae pmra in complex with pmra2 box dna
|
|
|
| 103 | c4ldaF_ |
|
not modelled |
90.4 |
13 |
PDB header:transcription
Chain: F: PDB Molecule:tadz;
PDBTitle: crystal structure of a chey-like protein (tadz) from pseudomonas2 aeruginosa pao1 at 2.70 a resolution
|
|
|
| 104 | c4z87B_ |
|
not modelled |
90.3 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: structure of the imp dehydrogenase from ashbya gossypii bound to gdp
|
|
|
| 105 | c5m7nA_ |
|
not modelled |
89.9 |
13 |
PDB header:signaling protein
Chain: A: PDB Molecule:nitrogen assimilation regulatory protein;
PDBTitle: crystal structure of ntrx from brucella abortus in complex with atp2 processed with the crystaldirect automated mounting and cryo-cooling3 technology
|
|
|
| 106 | c3c3mA_ |
|
not modelled |
89.9 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of the n-terminal domain of response regulator2 receiver protein from methanoculleus marisnigri jr1
|
|
|
| 107 | d1zgza1 |
|
not modelled |
89.8 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 108 | c3jteA_ |
|
not modelled |
89.7 |
11 |
PDB header:protein binding
Chain: A: PDB Molecule:response regulator receiver protein;
PDBTitle: crystal structure of response regulator receiver domain protein from2 clostridium thermocellum
|
|
|
| 109 | c2gwrA_ |
|
not modelled |
89.6 |
12 |
PDB header:signaling protein
Chain: A: PDB Molecule:dna-binding response regulator mtra;
PDBTitle: crystal structure of the response regulator protein mtra from2 mycobacterium tuberculosis
|
|
|
| 110 | c3a0rB_ |
|
not modelled |
89.6 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:response regulator;
PDBTitle: crystal structure of histidine kinase thka (tm1359) in complex with2 response regulator protein trra (tm1360)
|
|
|
| 111 | c3eq2A_ |
|
not modelled |
89.6 |
20 |
PDB header:signaling protein
Chain: A: PDB Molecule:probable two-component response regulator;
PDBTitle: structure of hexagonal crystal form of pseudomonas aeruginosa rssb
|
|
|
| 112 | d1jbea_ |
|
not modelled |
89.2 |
16 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 113 | c1w25B_ |
|
not modelled |
88.9 |
8 |
PDB header:signaling protein
Chain: B: PDB Molecule:stalked-cell differentiation controlling protein;
PDBTitle: response regulator pled in complex with c-digmp
|
|
|
| 114 | d1yx1a1 |
|
not modelled |
88.9 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:KguE-like |
|
|
| 115 | c3ivuB_ |
|
not modelled |
88.6 |
16 |
PDB header:transferase
Chain: B: PDB Molecule:homocitrate synthase, mitochondrial;
PDBTitle: homocitrate synthase lys4 bound to 2-og
|
|
|
| 116 | c4fxsA_ |
|
not modelled |
88.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine-5'-monophosphate dehydrogenase;
PDBTitle: inosine 5'-monophosphate dehydrogenase from vibrio cholerae complexed2 with imp and mycophenolic acid
|
|
|
| 117 | c3cu5B_ |
|
not modelled |
88.4 |
8 |
PDB header:transcription regulator
Chain: B: PDB Molecule:two component transcriptional regulator, arac family;
PDBTitle: crystal structure of a two component transcriptional regulator arac2 from clostridium phytofermentans isdg
|
|
|
| 118 | c3r0jA_ |
|
not modelled |
88.2 |
15 |
PDB header:dna binding protein
Chain: A: PDB Molecule:possible two component system response transcriptional
PDBTitle: structure of phop from mycobacterium tuberculosis
|
|
|
| 119 | c3fd0B_ |
|
not modelled |
87.9 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:putative cystathionine beta-lyase involved in aluminum
PDBTitle: crystal structure of putative cystathionine beta-lyase involved in2 aluminum resistance (np_470671.1) from listeria innocua at 2.12 a3 resolution
|
|
|
| 120 | c6ekhY_ |
|
not modelled |
87.9 |
13 |
PDB header:metal binding protein
Chain: Y: PDB Molecule:chemotaxis protein chey;
PDBTitle: crystal structure of activated chey from methanoccocus maripaludis
|
|
|