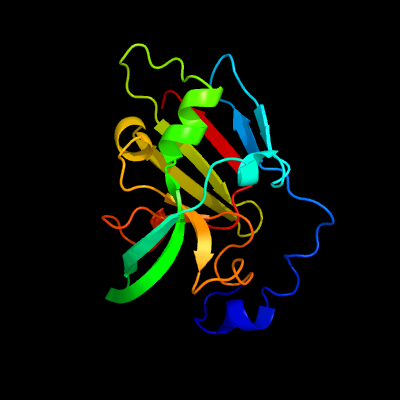
| 1 | d1xdya_
|
|
 |
100.0 |
28 |
Fold:Oxidoreductase molybdopterin-binding domain
Superfamily:Oxidoreductase molybdopterin-binding domain
Family:Oxidoreductase molybdopterin-binding domain |
|
|
|
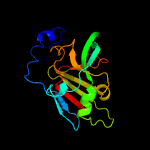
| 2 | c1xdyC_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:bacterial sulfite oxidase;
PDBTitle: structural and biochemical identification of a novel2 bacterial oxidoreductase, w-containing cofactor
|
|
|
|
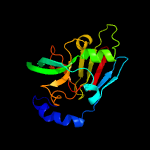
| 3 | c2xtsC_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase/electron transport
Chain: C: PDB Molecule:sulfite dehydrogenase;
PDBTitle: crystal structure of the sulfane dehydrogenase soxcd from paracoccus2 pantotrophus
|
|
|
|
| 4 | c1soxB_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sulfite oxidase;
PDBTitle: sulfite oxidase from chicken liver
|
|
|
|
| 5 | c2bpbA_
|
|
 |
100.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfite\:cytochrome c oxidoreductase subunit a;
PDBTitle: sulfite dehydrogenase from starkeya novella
|
|
|
|
| 6 | c2bihA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitrate reductase [nadph];
PDBTitle: crystal structure of the molybdenum-containing nitrate2 reducing fragment of pichia angusta assimilatory nitrate3 reductase
|
|
|
|
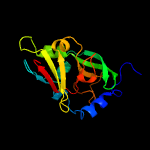
| 7 | c1ogpD_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:sulfite oxidase;
PDBTitle: the crystal structure of plant sulfite oxidase provides2 insight into sulfite oxidation in plants and animals
|
|
|
|
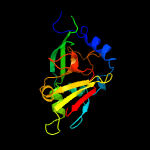
| 8 | d1ogpa2
|
|
 |
100.0 |
27 |
Fold:Oxidoreductase molybdopterin-binding domain
Superfamily:Oxidoreductase molybdopterin-binding domain
Family:Oxidoreductase molybdopterin-binding domain |
|
|
|
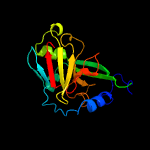
| 9 | c2a9dB_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:sulfite oxidase;
PDBTitle: crystal structure of recombinant chicken sulfite oxidase with arg at2 residue 161
|
|
|
|
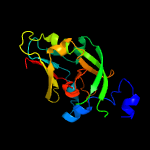
| 10 | c2biiA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitrate reductase [nadph];
PDBTitle: crystal structure of nitrate-reducing fragment of2 assimilatory nitrate reductase from pichia angusta
|
|
|
|
| 11 | d2a9da2
|
|
 |
100.0 |
27 |
Fold:Oxidoreductase molybdopterin-binding domain
Superfamily:Oxidoreductase molybdopterin-binding domain
Family:Oxidoreductase molybdopterin-binding domain |
|
|
|
| 12 | c4pw3B_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative sulfite oxidase;
PDBTitle: crystal structure of the sulfite dehydrogenase sort from sinorhizobium2 meliloti
|
|
|
|
| 13 | d1kqfc_
|
|
 |
98.4 |
15 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Formate dehydrogenase N, cytochrome (gamma) subunit |
|
|
|
| 14 | c4gd3A_
|
|
 |
97.9 |
21 |
PDB header:oxidoreductase/electron transport
Chain: A: PDB Molecule:ni/fe-hydrogenase 1 b-type cytochrome subunit;
PDBTitle: structure of e. coli hydrogenase-1 in complex with cytochrome b
|
|
|
|
| 15 | c4gd3B_
|
|
 |
97.9 |
21 |
PDB header:oxidoreductase/electron transport
Chain: B: PDB Molecule:ni/fe-hydrogenase 1 b-type cytochrome subunit;
PDBTitle: structure of e. coli hydrogenase-1 in complex with cytochrome b
|
|
|
|
| 16 | c5oc0A_
|
|
 |
96.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome b561;
PDBTitle: structure of e. coli superoxide oxidase
|
|
|
|
| 17 | c6hwhb_
|
|
 |
95.0 |
20 |
PDB header:electron transport
Chain: B: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
|
|
|
|
| 18 | c2qjkM_
|
|
 |
94.5 |
20 |
PDB header:electron transport
Chain: M: PDB Molecule:cytochrome b;
PDBTitle: crystal structure analysis of mutant rhodobacter2 sphaeroides bc1 with stigmatellin and antimycin
|
|
|
|
| 19 | d1q90b_
|
|
 |
92.9 |
19 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
|
|
|
| 20 | d2e74a1
|
|
 |
91.7 |
19 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
|
|
|
| 21 | d1ppjc2 |
|
not modelled |
89.6 |
17 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
|
|
| 22 | c3cwbC_ |
|
not modelled |
89.1 |
20 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:cytochrome b;
PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
|
|
|
| 23 | c3cx5N_ |
|
not modelled |
88.9 |
15 |
PDB header:oxidoreductase
Chain: N: PDB Molecule:cytochrome b;
PDBTitle: structure of complex iii with bound cytochrome c in reduced state and2 definition of a minimal core interface for electron transfer.
|
|
|
| 24 | d3cx5c2 |
|
not modelled |
87.7 |
15 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
|
|
| 25 | d1bccc3 |
|
not modelled |
81.2 |
20 |
Fold:Heme-binding four-helical bundle
Superfamily:Transmembrane di-heme cytochromes
Family:Cytochrome b of cytochrome bc1 complex (Ubiquinol-cytochrome c reductase) |
|
|
| 26 | c6f0kC_ |
|
not modelled |
72.8 |
16 |
PDB header:membrane protein
Chain: C: PDB Molecule:polysulphide reductase nrfd;
PDBTitle: alternative complex iii
|
|
|
| 27 | c2e76D_ |
|
not modelled |
61.8 |
14 |
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
|
|
| 28 | c6hwhB_ |
|
not modelled |
50.4 |
21 |
PDB header:electron transport
Chain: B: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
|
|
|
| 29 | c6btmC_ |
|
not modelled |
39.4 |
18 |
PDB header:membrane protein
Chain: C: PDB Molecule:alternative complex iii subunit c;
PDBTitle: structure of alternative complex iii from flavobacterium johnsoniae2 (wild type)
|
|
|
| 30 | d1nkza_ |
|
not modelled |
34.3 |
24 |
Fold:Light-harvesting complex subunits
Superfamily:Light-harvesting complex subunits
Family:Light-harvesting complex subunits |
|
|
| 31 | d1ny721 |
|
not modelled |
27.7 |
26 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Comoviridae-like VP |
|
|
| 32 | d1pgw21 |
|
not modelled |
27.0 |
26 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Comoviridae-like VP |
|
|
| 33 | d1pgl21 |
|
not modelled |
25.9 |
26 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Comoviridae-like VP |
|
|
| 34 | c2fyuE_ |
|
not modelled |
24.1 |
23 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
|
|
| 35 | c5npxL_ |
|
not modelled |
23.7 |
29 |
PDB header:virus
Chain: L: PDB Molecule:polyprotein;
PDBTitle: atomic structure of the broad bean stain virus (bbsv) by cryo-em
|
|
|
| 36 | c4o2tB_ |
|
not modelled |
22.0 |
29 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a duf4827 family protein (bdi_1692) from2 parabacteroides distasonis atcc 8503 at 2.40 a resolution
|
|
|
| 37 | d2axta1 |
|
not modelled |
21.8 |
29 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 38 | d1ijda_ |
|
not modelled |
20.7 |
25 |
Fold:Light-harvesting complex subunits
Superfamily:Light-harvesting complex subunits
Family:Light-harvesting complex subunits |
|
|
| 39 | c1bmv2_ |
|
not modelled |
19.8 |
25 |
PDB header:virus/rna
Chain: 2: PDB Molecule:protein (icosahedral virus - b and c domain);
PDBTitle: protein-rna interactions in an icosahedral virus at 3.02 angstroms resolution
|
|
|
| 40 | c1vw4b_ |
|
not modelled |
19.1 |
27 |
PDB header:ribosome
Chain: B: PDB Molecule:
PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
|
|
|
| 41 | c2fynO_ |
|
not modelled |
17.7 |
24 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
|
|
| 42 | d1h9aa2 |
|
not modelled |
15.0 |
18 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:Glucose 6-phosphate dehydrogenase-like |
|
|
| 43 | d2evea1 |
|
not modelled |
13.1 |
23 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Atu2648/PH1033-like |
|
|
| 44 | c6igzH_ |
|
not modelled |
11.5 |
20 |
PDB header:plant protein
Chain: H: PDB Molecule:psah;
PDBTitle: structure of psi-lhci
|
|
|
| 45 | c4e9iB_ |
|
not modelled |
11.3 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: glucose-6-p dehydrogenase (apo form) from trypanosoma cruzi
|
|
|
| 46 | c2ky9A_ |
|
not modelled |
10.8 |
40 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein ydhk;
PDBTitle: solution nmr structure of ydhk c-terminal domain from b.subtilis,2 northeast structural genomics consortium target target sr518
|
|
|
| 47 | d2g2xa1 |
|
not modelled |
9.9 |
36 |
Fold:PUA domain-like
Superfamily:PUA domain-like
Family:Atu2648/PH1033-like |
|
|
| 48 | c1h9aA_ |
|
not modelled |
9.2 |
18 |
PDB header:oxidoreductase (choh(d) - nad(p))
Chain: A: PDB Molecule:glucose 6-phosphate 1-dehydrogenase;
PDBTitle: complex of active mutant (q365->c) of glucose 6-phosphate2 dehydrogenase from l. mesenteroides with coenzyme nadp
|
|
|
| 49 | c4lgvA_ |
|
not modelled |
8.8 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray crystal structure of glucose-6-phosphate 1-dehydrogenase from2 mycobacterium avium
|
|
|
| 50 | c2bfuL_ |
|
not modelled |
8.7 |
26 |
PDB header:virus
Chain: L: PDB Molecule:cowpea mosaic virus, large (l) subunit;
PDBTitle: x-ray structure of cpmv top component
|
|
|
| 51 | c2bhlB_ |
|
not modelled |
8.4 |
27 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray structure of human glucose-6-phosphate dehydrogenase (deletion2 variant) complexed with glucose-6-phosphate
|
|
|
| 52 | c1qkiE_ |
|
not modelled |
8.3 |
21 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray structure of human glucose 6-phosphate dehydrogenase (variant2 canton r459l) complexed with structural nadp+
|
|
|
| 53 | c1p84E_ |
|
not modelled |
8.1 |
9 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
|
|
| 54 | c1ej6D_ |
|
not modelled |
7.9 |
13 |
PDB header:virus
Chain: D: PDB Molecule:sigma2;
PDBTitle: reovirus core
|
|
|
| 55 | c3diiB_ |
|
not modelled |
7.7 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of a carbohydrate specific scor enzyme2 from clostridium thermocellum, ligand-free form
|
|
|
| 56 | d1qkia2 |
|
not modelled |
7.7 |
21 |
Fold:FwdE/GAPDH domain-like
Superfamily:Glyceraldehyde-3-phosphate dehydrogenase-like, C-terminal domain
Family:Glucose 6-phosphate dehydrogenase-like |
|
|
| 57 | c2o01H_ |
|
not modelled |
7.7 |
14 |
PDB header:photosynthesis
Chain: H: PDB Molecule:photosystem i reaction center subunit vi, chloroplast;
PDBTitle: the structure of a plant photosystem i supercomplex at 3.4 angstrom2 resolution
|
|
|
| 58 | c6fo2R_ |
|
not modelled |
7.6 |
16 |
PDB header:membrane protein
Chain: R: PDB Molecule:cytochrome b-c1 complex subunit rieske, mitochondrial;
PDBTitle: cryoem structure of bovine cytochrome bc1 with no ligand bound
|
|
|
| 59 | c2pq4B_ |
|
not modelled |
6.9 |
24 |
PDB header:chaperone/oxidoreductase
Chain: B: PDB Molecule:periplasmic nitrate reductase precursor;
PDBTitle: nmr solution structure of napd in complex with napa1-352 signal peptide
|
|
|
| 60 | c5zjiH_ |
|
not modelled |
6.6 |
12 |
PDB header:membrane protein
Chain: H: PDB Molecule:photosystem i reaction center subunit vi, chloroplastic;
PDBTitle: structure of photosystem i supercomplex with light-harvesting2 complexes i and ii
|
|
|
| 61 | c5ghaF_ |
|
not modelled |
6.1 |
33 |
PDB header:transferase/transport protein
Chain: F: PDB Molecule:sulfur carrier ttub;
PDBTitle: sulfur transferase ttua in complex with sulfur carrier ttub
|
|
|
| 62 | d1r6oc1 |
|
not modelled |
6.1 |
9 |
Fold:ClpS-like
Superfamily:ClpS-like
Family:Adaptor protein ClpS (YljA) |
|
|
| 63 | c2ol5B_ |
|
not modelled |
6.0 |
7 |
PDB header:transcription regulator
Chain: B: PDB Molecule:pai 2 protein;
PDBTitle: crystal structure of a protease synthase and sporulation negative2 regulatory protein pai 2 from bacillus stearothermophilus
|
|
|
| 64 | c5ykaA_ |
|
not modelled |
5.9 |
18 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:uncharacterized protein kdoo;
PDBTitle: crystal structure of the kdo hydroxylase kdoo, a non-heme fe(ii)2 alphaketoglutarate dependent dioxygenase in complex with cobalt(ii)
|
|
|
| 65 | c3llkA_ |
|
not modelled |
5.5 |
35 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:sulfhydryl oxidase 1;
PDBTitle: sulfhydryl oxidase fragment of human qsox1
|
|
|
| 66 | d1f46a_ |
|
not modelled |
5.1 |
13 |
Fold:TBP-like
Superfamily:Cell-division protein ZipA, C-terminal domain
Family:Cell-division protein ZipA, C-terminal domain |
|
|
| 67 | d2i5nl1 |
|
not modelled |
5.1 |
23 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|