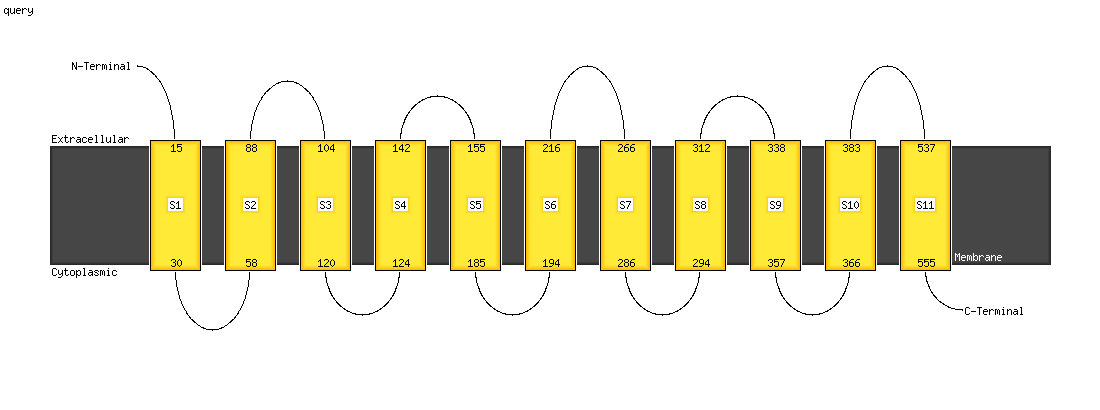
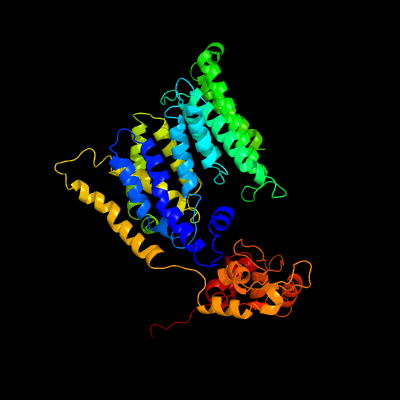
| 1 | c3wajA_
|
|
 |
99.1 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:transmembrane oligosaccharyl transferase;
PDBTitle: crystal structure of the archaeoglobus fulgidus2 oligosaccharyltransferase (o29867_arcfu) complex with zn and sulfate
|
|
|
|
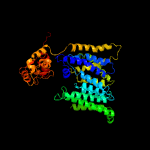
| 2 | c3rceA_
|
|
 |
98.6 |
11 |
PDB header:transferase/peptide
Chain: A: PDB Molecule:oligosaccharide transferase to n-glycosylate proteins;
PDBTitle: bacterial oligosaccharyltransferase pglb
|
|
|
|
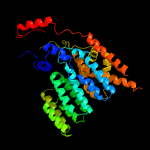
| 3 | c6eznF_
|
|
 |
98.4 |
12 |
PDB header:membrane protein
Chain: F: PDB Molecule:dolichyl-diphosphooligosaccharide--protein
PDBTitle: cryo-em structure of the yeast oligosaccharyltransferase (ost) complex
|
|
|
|
| 4 | c5f15A_
|
|
 |
97.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:4-amino-4-deoxy-l-arabinose (l-ara4n) transferase;
PDBTitle: crystal structure of arnt from cupriavidus metallidurans bound to2 undecaprenyl phosphate
|
|
|
|
| 5 | c3vu0B_
|
|
 |
33.6 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (afaglb-s2, af_0040, o30195_arcfu) from3 archaeoglobus fulgidus
|
|
|
|
| 6 | c3vgpA_
|
|
 |
25.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:transmembrane oligosaccharyl transferase, putative;
PDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (af_0329) from archaeoglobus fulgidus
|
|
|
|
| 7 | c2l9uA_
|
|
 |
25.3 |
42 |
PDB header:membrane protein
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-3;
PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
|
|
|
|
| 8 | c3waiA_
|
|
 |
21.9 |
10 |
PDB header:transferase, transport protein
Chain: A: PDB Molecule:maltose-binding periplasmic protein, transmembrane
PDBTitle: crystal structure of the c-terminal globular domain of2 oligosaccharyltransferase (afaglb-l, o29867_arcfu) from archaeoglobus3 fulgidus as a mbp fusion
|
|
|
|
| 9 | c4dl0I_
|
|
 |
21.1 |
28 |
PDB header:hydrolase
Chain: I: PDB Molecule:v-type proton atpase subunit c;
PDBTitle: crystal structure of the heterotrimeric egchead peripheral stalk2 complex of the yeast vacuolar atpase
|
|
|
|
| 10 | d1qkia1
|
|
 |
19.9 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
|
| 11 | c6p25A_
|
|
 |
18.7 |
5 |
PDB header:transferase
Chain: A: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 1;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor and a peptide acceptor
|
|
|
|
| 12 | d1h9aa1
|
|
 |
18.1 |
26 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Glyceraldehyde-3-phosphate dehydrogenase-like, N-terminal domain |
|
|
|
| 13 | c5vylA_
|
|
 |
17.9 |
37 |
PDB header:viral protein
Chain: A: PDB Molecule:inner tegument protein;
PDBTitle: crystal structure of n-terminal half of herpes simplex virus type 12 ul37 protein
|
|
|
|
| 14 | c4k70B_
|
|
 |
17.1 |
25 |
PDB header:viral protein
Chain: B: PDB Molecule:ul37;
PDBTitle: crystal structure of n-terminal half of pseudorabiesvirus ul37 protein
|
|
|
|
| 15 | d1a77a2
|
|
 |
13.1 |
19 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
|
| 16 | c3gxgA_
|
|
 |
12.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative phosphatase (duf442);
PDBTitle: crystal structure of putative phosphatase (duf442) (yp_001181608.1)2 from shewanella putrefaciens cn-32 at 1.60 a resolution
|
|
|
|
| 17 | c4j2lC_
|
|
 |
12.0 |
40 |
PDB header:transcription regulator
Chain: C: PDB Molecule:protein capicua homolog;
PDBTitle: crystal structure of axh domain complexed with capicua
|
|
|
|
| 18 | c4j2lD_
|
|
 |
12.0 |
40 |
PDB header:transcription regulator
Chain: D: PDB Molecule:protein capicua homolog;
PDBTitle: crystal structure of axh domain complexed with capicua
|
|
|
|
| 19 | c6p2rB_
|
|
 |
11.7 |
7 |
PDB header:transferase
Chain: B: PDB Molecule:dolichyl-phosphate-mannose--protein mannosyltransferase 2;
PDBTitle: structure of s. cerevisiae protein o-mannosyltransferase pmt1-pmt22 complex bound to the sugar donor
|
|
|
|
| 20 | c5kzoA_
|
|
 |
11.6 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:neurogenic locus notch homolog protein 1;
PDBTitle: notch1 transmembrane and associated juxtamembrane segment
|
|
|
|
| 21 | d1u7la_ |
|
not modelled |
11.5 |
28 |
Fold:Vacuolar ATP synthase subunit C
Superfamily:Vacuolar ATP synthase subunit C
Family:Vacuolar ATP synthase subunit C |
|
|
| 22 | d2i5nm1 |
|
not modelled |
11.2 |
18 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 23 | c2b5dX_ |
|
not modelled |
10.5 |
21 |
PDB header:hydrolase
Chain: X: PDB Molecule:alpha-amylase;
PDBTitle: crystal structure of the novel alpha-amylase amyc from thermotoga2 maritima
|
|
|
| 24 | c2f46A_ |
|
not modelled |
10.2 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:hypothetical protein;
PDBTitle: crystal structure of a putative phosphatase (nma1982) from neisseria2 meningitidis z2491 at 1.41 a resolution
|
|
|
| 25 | d1rxwa2 |
|
not modelled |
10.1 |
13 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
| 26 | d1eysl_ |
|
not modelled |
9.7 |
11 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 27 | d1vkia_ |
|
not modelled |
9.7 |
6 |
Fold:YbaK/ProRS associated domain
Superfamily:YbaK/ProRS associated domain
Family:YbaK/ProRS associated domain |
|
|
| 28 | d1eysm_ |
|
not modelled |
9.6 |
13 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 29 | d2axtd1 |
|
not modelled |
9.4 |
14 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 30 | c2m7yA_ |
|
not modelled |
9.4 |
29 |
PDB header:viral protein
Chain: A: PDB Molecule:leader peptide;
PDBTitle: the mengovirus leader protein
|
|
|
| 31 | d2j8cm1 |
|
not modelled |
9.3 |
15 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 32 | d2i5nl1 |
|
not modelled |
9.3 |
13 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 33 | d1u6ka1 |
|
not modelled |
9.2 |
22 |
Fold:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Superfamily:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD)
Family:F420-dependent methylenetetrahydromethanopterin dehydrogenase (MTD) |
|
|
| 34 | c4witB_ |
|
not modelled |
9.2 |
20 |
PDB header:lipid transport
Chain: B: PDB Molecule:predicted protein;
PDBTitle: tmem16 lipid scramblase in crystal form 2
|
|
|
| 35 | c1k1yA_ |
|
not modelled |
9.1 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:4-alpha-glucanotransferase;
PDBTitle: crystal structure of thermococcus litoralis 4-alpha-glucanotransferase2 complexed with acarbose
|
|
|
| 36 | d2b5dx2 |
|
not modelled |
9.0 |
21 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:AmyC N-terminal domain-like |
|
|
| 37 | c3n92A_ |
|
not modelled |
8.9 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:alpha-amylase, gh57 family;
PDBTitle: crystal structure of tk1436, a gh57 branching enzyme from2 hyperthermophilic archaeon thermococcus kodakaraensis, in complex3 with glucose
|
|
|
| 38 | d2pp6a1 |
|
not modelled |
8.8 |
22 |
Fold:Phage tail proteins
Superfamily:Phage tail proteins
Family:gpFII-like |
|
|
| 39 | d2j8cl1 |
|
not modelled |
8.8 |
10 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 40 | d1l9bm_ |
|
not modelled |
8.8 |
14 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 41 | c2lowA_ |
|
not modelled |
8.7 |
13 |
PDB header:membrane protein
Chain: A: PDB Molecule:apelin receptor;
PDBTitle: solution structure of ar55 in 50% hfip
|
|
|
| 42 | d2axta1 |
|
not modelled |
8.6 |
13 |
Fold:Bacterial photosystem II reaction centre, L and M subunits
Superfamily:Bacterial photosystem II reaction centre, L and M subunits
Family:Bacterial photosystem II reaction centre, L and M subunits |
|
|
| 43 | c5yq7M_ |
|
not modelled |
8.2 |
14 |
PDB header:photosynthesis
Chain: M: PDB Molecule:precursor for m subunits of photosynthetic reaction center;
PDBTitle: cryo-em structure of the rc-lh core complex from roseiflexus2 castenholzii
|
|
|
| 44 | c5yq7L_ |
|
not modelled |
8.1 |
6 |
PDB header:photosynthesis
Chain: L: PDB Molecule:precursor for l subunits of photosynthetic reaction center;
PDBTitle: cryo-em structure of the rc-lh core complex from roseiflexus2 castenholzii
|
|
|
| 45 | c2bhlB_ |
|
not modelled |
8.0 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray structure of human glucose-6-phosphate dehydrogenase (deletion2 variant) complexed with glucose-6-phosphate
|
|
|
| 46 | c2n1pA_ |
|
not modelled |
8.0 |
38 |
PDB header:viral protein
Chain: A: PDB Molecule:non-structural protein 5b, ns5b;
PDBTitle: structure of the c-terminal membrane domain of hcv ns5b protein
|
|
|
| 47 | c6g90J_ |
|
not modelled |
7.9 |
50 |
PDB header:splicing
Chain: J: PDB Molecule:u1 small nuclear ribonucleoprotein component snu71;
PDBTitle: prespliceosome structure provides insight into spliceosome assembly2 and regulation (map a2)
|
|
|
| 48 | d1m1ha2 |
|
not modelled |
7.8 |
6 |
Fold:Ferredoxin-like
Superfamily:N-utilization substance G protein NusG, N-terminal domain
Family:N-utilization substance G protein NusG, N-terminal domain |
|
|
| 49 | d1tu7a2 |
|
not modelled |
7.7 |
5 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
|
|
| 50 | c1qkiE_ |
|
not modelled |
7.4 |
24 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray structure of human glucose 6-phosphate dehydrogenase (variant2 canton r459l) complexed with structural nadp+
|
|
|
| 51 | d1ufaa2 |
|
not modelled |
7.3 |
21 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:AmyC N-terminal domain-like |
|
|
| 52 | c1a77A_ |
|
not modelled |
7.3 |
19 |
PDB header:5'-3' exo/endo nuclease
Chain: A: PDB Molecule:flap endonuclease-1 protein;
PDBTitle: flap endonuclease-1 from methanococcus jannaschii
|
|
|
| 53 | d2cvda2 |
|
not modelled |
7.2 |
12 |
Fold:Thioredoxin fold
Superfamily:Thioredoxin-like
Family:Glutathione S-transferase (GST), N-terminal domain |
|
|
| 54 | d1k1xa3 |
|
not modelled |
7.2 |
26 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:4-alpha-glucanotransferase, N-terminal domain |
|
|
| 55 | c1ufaA_ |
|
not modelled |
7.2 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:tt1467 protein;
PDBTitle: crystal structure of tt1467 from thermus thermophilus hb8
|
|
|
| 56 | c1h9aA_ |
|
not modelled |
6.8 |
26 |
PDB header:oxidoreductase (choh(d) - nad(p))
Chain: A: PDB Molecule:glucose 6-phosphate 1-dehydrogenase;
PDBTitle: complex of active mutant (q365->c) of glucose 6-phosphate2 dehydrogenase from l. mesenteroides with coenzyme nadp
|
|
|
| 57 | c5n6mA_ |
|
not modelled |
6.8 |
16 |
PDB header:membrane protein
Chain: A: PDB Molecule:apolipoprotein n-acyltransferase;
PDBTitle: structure of the membrane integral lipoprotein n-acyltransferase lnt2 from p. aeruginosa
|
|
|
| 58 | d1m6ja_ |
|
not modelled |
6.6 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 59 | c3qeaZ_ |
|
not modelled |
6.4 |
16 |
PDB header:hydrolase/dna
Chain: Z: PDB Molecule:exonuclease 1;
PDBTitle: crystal structure of human exonuclease 1 exo1 (wt) in complex with dna2 (complex ii)
|
|
|
| 60 | c4y8fA_ |
|
not modelled |
6.4 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from clostridium2 perfringens
|
|
|
| 61 | c1b43A_ |
|
not modelled |
6.3 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:protein (fen-1);
PDBTitle: fen-1 from p. furiosus
|
|
|
| 62 | c3s6dA_ |
|
not modelled |
6.2 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:putative triosephosphate isomerase;
PDBTitle: crystal structure of a putative triosephosphate isomerase from2 coccidioides immitis
|
|
|
| 63 | c4y9aB_ |
|
not modelled |
6.2 |
20 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from streptomyces2 coelicolor
|
|
|
| 64 | d2j07a2 |
|
not modelled |
6.2 |
21 |
Fold:Cryptochrome/photolyase, N-terminal domain
Superfamily:Cryptochrome/photolyase, N-terminal domain
Family:Cryptochrome/photolyase, N-terminal domain |
|
|
| 65 | d1b43a2 |
|
not modelled |
6.1 |
19 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
| 66 | c5nc8B_ |
|
not modelled |
6.1 |
18 |
PDB header:transport protein
Chain: B: PDB Molecule:potassium efflux system protein;
PDBTitle: shewanella denitrificans kef ctd in amp bound form
|
|
|
| 67 | c4mknA_ |
|
not modelled |
6.1 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of chloroplastic triosephosphate isomerase from2 chlamydomonas reinhardtii at 1.1 a of resolution
|
|
|
| 68 | c4lgvA_ |
|
not modelled |
6.0 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: x-ray crystal structure of glucose-6-phosphate 1-dehydrogenase from2 mycobacterium avium
|
|
|
| 69 | d2beia1 |
|
not modelled |
5.9 |
4 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 70 | d1ul1x2 |
|
not modelled |
5.9 |
21 |
Fold:PIN domain-like
Superfamily:PIN domain-like
Family:5' to 3' exonuclease catalytic domain |
|
|
| 71 | c3op6B_ |
|
not modelled |
5.8 |
14 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of an oligo-nucleotide binding protein (lpg1207)2 from legionella pneumophila subsp. pneumophila str. philadelphia 1 at3 2.00 a resolution
|
|
|
| 72 | c5zg5B_ |
|
not modelled |
5.8 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase sadsubaaa mutant from2 opisthorchis viverrini
|
|
|
| 73 | c2jgqB_ |
|
not modelled |
5.8 |
12 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: kinetics and structural properties of triosephosphate2 isomerase from helicobacter pylori
|
|
|
| 74 | c5ujwD_ |
|
not modelled |
5.8 |
18 |
PDB header:isomerase
Chain: D: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from francisella2 tularensis subsp. tularensis schu s4
|
|
|
| 75 | c2mkvA_ |
|
not modelled |
5.8 |
13 |
PDB header:transport protein
Chain: A: PDB Molecule:sodium/potassium-transporting atpase subunit gamma;
PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
|
|
|
| 76 | c2ct6A_ |
|
not modelled |
5.8 |
10 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:sh3 domain-binding glutamic acid-rich-like
PDBTitle: solution structure of the sh3 domain-binding glutamic acid-2 rich-like protein 2
|
|
|
| 77 | c3oryA_ |
|
not modelled |
5.8 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:flap endonuclease 1;
PDBTitle: crystal structure of flap endonuclease 1 from hyperthermophilic2 archaeon desulfurococcus amylolyticus
|
|
|
| 78 | c2l9uB_ |
|
not modelled |
5.8 |
24 |
PDB header:membrane protein
Chain: B: PDB Molecule:receptor tyrosine-protein kinase erbb-3;
PDBTitle: spatial structure of dimeric erbb3 transmembrane domain
|
|
|
| 79 | c5wsnD_ |
|
not modelled |
5.7 |
16 |
PDB header:virus
Chain: D: PDB Molecule:m protein;
PDBTitle: structure of japanese encephalitis virus
|
|
|
| 80 | c3krsB_ |
|
not modelled |
5.7 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: structure of triosephosphate isomerase from cryptosporidium parvum at2 1.55a resolution
|
|
|
| 81 | c3th6B_ |
|
not modelled |
5.6 |
18 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from rhipicephalus2 (boophilus) microplus.
|
|
|
| 82 | c4e9iB_ |
|
not modelled |
5.6 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glucose-6-phosphate 1-dehydrogenase;
PDBTitle: glucose-6-p dehydrogenase (apo form) from trypanosoma cruzi
|
|
|
| 83 | d1o5xa_ |
|
not modelled |
5.6 |
0 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 84 | c3j2pD_ |
|
not modelled |
5.5 |
16 |
PDB header:viral protein
Chain: D: PDB Molecule:small envelope protein m;
PDBTitle: cryoem structure of dengue virus envelope protein heterotetramer
|
|
|
| 85 | c4nvtD_ |
|
not modelled |
5.5 |
24 |
PDB header:isomerase
Chain: D: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from brucella2 melitensis
|
|
|
| 86 | c4y90B_ |
|
not modelled |
5.5 |
29 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from deinococcus2 radiodurans
|
|
|
| 87 | c5ibxB_ |
|
not modelled |
5.5 |
24 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: 1.65 angstrom crystal structure of triosephosphate isomerase (tim)2 from streptococcus pneumoniae
|
|
|
| 88 | d2ozua1 |
|
not modelled |
5.4 |
20 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:N-acetyl transferase, NAT |
|
|
| 89 | d1b9ba_ |
|
not modelled |
5.4 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 90 | d2ctda2 |
|
not modelled |
5.3 |
50 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
|
|
| 91 | c3m9yB_ |
|
not modelled |
5.3 |
35 |
PDB header:isomerase
Chain: B: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of triosephosphate isomerase from methicillin2 resistant staphylococcus aureus at 1.9 angstrom resolution
|
|
|
| 92 | c5uprA_ |
|
not modelled |
5.3 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: x-ray structure of a putative triosephosphate isomerase from2 toxoplasma gondii me49
|
|
|
| 93 | d1sw3a_ |
|
not modelled |
5.3 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 94 | c1yyaA_ |
|
not modelled |
5.3 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:triosephosphate isomerase;
PDBTitle: crystal structure of tt0473, putative triosephosphate isomerase from2 thermus thermophilus hb8
|
|
|
| 95 | c5ireD_ |
|
not modelled |
5.3 |
22 |
PDB header:virus
Chain: D: PDB Molecule:m protein;
PDBTitle: the cryo-em structure of zika virus
|
|
|
| 96 | c1p58F_ |
|
not modelled |
5.2 |
13 |
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
|
|
|
| 97 | c1p58E_ |
|
not modelled |
5.2 |
13 |
PDB header:virus
Chain: E: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
|
|
|
| 98 | d1aw1a_ |
|
not modelled |
5.2 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Triosephosphate isomerase (TIM)
Family:Triosephosphate isomerase (TIM) |
|
|
| 99 | d1vpqa_ |
|
not modelled |
5.2 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:TM1631-like
Family:TM1631-like |
|
|