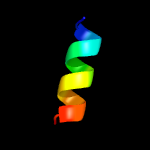| 1 |
|
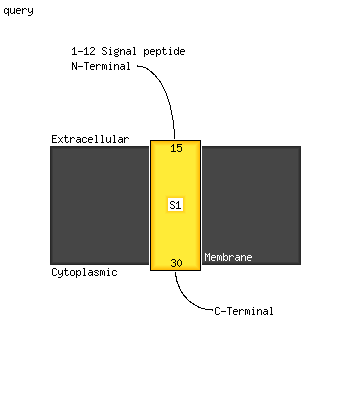
PDB 2k1l chain A
Region: 12 - 31
Aligned: 20
Modelled: 20
Confidence: 30.9%
Identity: 45%
PDB header:signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
Phyre2
| 2 |
|
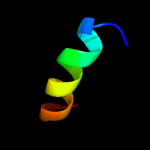
PDB 2k1k chain B
Region: 12 - 31
Aligned: 20
Modelled: 20
Confidence: 30.9%
Identity: 45%
PDB header:signaling protein
Chain: B: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
Phyre2
| 3 |
|
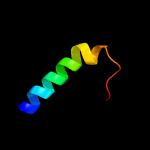
PDB 2k1k chain A
Region: 12 - 31
Aligned: 20
Modelled: 20
Confidence: 30.9%
Identity: 45%
PDB header:signaling protein
Chain: A: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 4.3
Phyre2
| 4 |
|
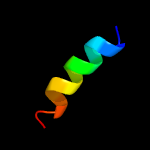
PDB 2k1l chain B
Region: 12 - 31
Aligned: 20
Modelled: 20
Confidence: 30.9%
Identity: 45%
PDB header:signaling protein
Chain: B: PDB Molecule:ephrin type-a receptor 1;
PDBTitle: nmr structures of dimeric transmembrane domain of the2 receptor tyrosine kinase epha1 in lipid bicelles at ph 6.3
Phyre2
| 5 |
|
PDB 1q90 chain R
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 17.0%
Identity: 42%
Fold: Single transmembrane helix
Superfamily: ISP transmembrane anchor
Family: ISP transmembrane anchor
Phyre2
| 6 |
|
PDB 1q90 chain R
Region: 1 - 24
Aligned: 24
Modelled: 24
Confidence: 17.0%
Identity: 42%
PDB header:photosynthesis
Chain: R: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: structure of the cytochrome b6f (plastohydroquinone : plastocyanin2 oxidoreductase) from chlamydomonas reinhardtii
Phyre2
| 7 |
|
PDB 4cdi chain C
Region: 4 - 36
Aligned: 33
Modelled: 33
Confidence: 16.9%
Identity: 24%
PDB header:membrane protein
Chain: C: PDB Molecule:predicted protein;
PDBTitle: crystal structure of acrb-acrz complex
Phyre2
| 8 |
|
PDB 2jwa chain A
Region: 11 - 34
Aligned: 24
Modelled: 24
Confidence: 15.3%
Identity: 42%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: erbb2 transmembrane segment dimer spatial structure
Phyre2
| 9 |
|
PDB 2ks1 chain A
Region: 11 - 34
Aligned: 24
Modelled: 24
Confidence: 15.3%
Identity: 42%
PDB header:transferase
Chain: A: PDB Molecule:receptor tyrosine-protein kinase erbb-2;
PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
Phyre2
| 10 |
|
PDB 4etr chain A
Region: 40 - 54
Aligned: 15
Modelled: 15
Confidence: 14.8%
Identity: 53%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: x-ray structure of pa2169 from pseudomonas aeruginosa
Phyre2
| 11 |
|
PDB 2lp1 chain A
Region: 11 - 26
Aligned: 16
Modelled: 16
Confidence: 11.2%
Identity: 31%
PDB header:membrane protein
Chain: A: PDB Molecule:c99;
PDBTitle: the solution nmr structure of the transmembrane c-terminal domain of2 the amyloid precursor protein (c99)
Phyre2
| 12 |
|
PDB 4xyp chain A
Region: 42 - 54
Aligned: 13
Modelled: 13
Confidence: 10.4%
Identity: 46%
PDB header:viral protein
Chain: A: PDB Molecule:fusion protein;
PDBTitle: crystal structure of a piscine viral fusion protein
Phyre2
| 13 |
|
PDB 1r7m chain A domain 1
Region: 5 - 21
Aligned: 17
Modelled: 17
Confidence: 9.0%
Identity: 29%
Fold: Homing endonuclease-like
Superfamily: Homing endonucleases
Family: Group I mobile intron endonuclease
Phyre2
| 14 |
|
PDB 2k9p chain A
Region: 12 - 38
Aligned: 27
Modelled: 27
Confidence: 8.7%
Identity: 26%
PDB header:membrane protein
Chain: A: PDB Molecule:pheromone alpha factor receptor;
PDBTitle: structure of tm1_tm2 in lppg micelles
Phyre2
| 15 |
|
PDB 1r7m chain A
Region: 5 - 21
Aligned: 17
Modelled: 17
Confidence: 7.7%
Identity: 29%
PDB header:hydrolase/dna
Chain: A: PDB Molecule:intron-encoded endonuclease i-scei;
PDBTitle: the homing endonuclease i-scei bound to its dna recognition2 region
Phyre2
| 16 |
|
PDB 1jb0 chain K
Region: 10 - 25
Aligned: 16
Modelled: 16
Confidence: 7.2%
Identity: 38%
PDB header:photosynthesis
Chain: K: PDB Molecule:photosystem 1 reaction centre subunit x;
PDBTitle: crystal structure of photosystem i: a photosynthetic reaction center2 and core antenna system from cyanobacteria
Phyre2
| 17 |
|
PDB 1jb0 chain K
Region: 10 - 25
Aligned: 16
Modelled: 16
Confidence: 7.2%
Identity: 38%
Fold: Photosystem I reaction center subunit X, PsaK
Superfamily: Photosystem I reaction center subunit X, PsaK
Family: Photosystem I reaction center subunit X, PsaK
Phyre2
| 18 |
|
PDB 3q7c chain A
Region: 6 - 17
Aligned: 12
Modelled: 12
Confidence: 6.6%
Identity: 33%
PDB header:hydrolase
Chain: A: PDB Molecule:nucleoprotein;
PDBTitle: exonuclease domain of lassa virus nucleoprotein bound to manganese
Phyre2
| 19 |
|
PDB 2k8p chain A
Region: 35 - 57
Aligned: 23
Modelled: 23
Confidence: 6.5%
Identity: 30%
PDB header:signaling protein
Chain: A: PDB Molecule:sclerostin;
PDBTitle: characterisation of the structural features and2 interactions of sclerostin: molecular insight into a key3 regulator of wnt-mediated bone formation
Phyre2
| 20 |
|
PDB 1h41 chain A domain 2
Region: 21 - 40
Aligned: 20
Modelled: 20
Confidence: 6.2%
Identity: 15%
Fold: Zincin-like
Superfamily: beta-N-acetylhexosaminidase-like domain
Family: alpha-D-glucuronidase, N-terminal domain
Phyre2
| 21 |
|
| 22 |
|
| 23 |
|