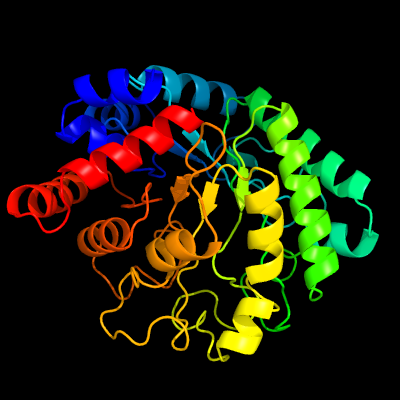
| 1 | c3bmxB_
|
|
 |
100.0 |
33 |
PDB header:hydrolase
Chain: B: PDB Molecule:uncharacterized lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase (ybbd) from bacillus subtilis
|
|
|
|
| 2 | c3sqlB_
|
|
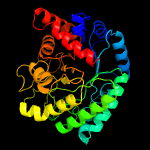 |
100.0 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase family 3;
PDBTitle: crystal structure of glycoside hydrolase from synechococcus
|
|
|
|
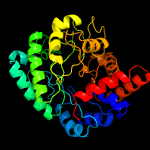
| 3 | c4yyfC_
|
|
 |
100.0 |
73 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: the crystal structure of a glycosyl hydrolase of gh3 family member2 from [mycobacterium smegmatis str. mc2 155
|
|
|
|
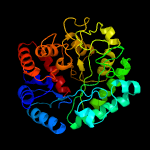
| 4 | c4zm6A_
|
|
 |
100.0 |
32 |
PDB header:hydrolase, transferase
Chain: A: PDB Molecule:n-acetyl-beta-d glucosaminidase;
PDBTitle: a unique gcn5-related glucosamine n-acetyltransferase region exist in2 the fungal multi-domain gh3 beta-n-acetylglucosaminidase
|
|
|
|
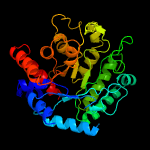
| 5 | c3lk6A_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:lipoprotein ybbd;
PDBTitle: beta-n-hexosaminidase n318d mutant (ybbd_n318d) from bacillus subtilis
|
|
|
|
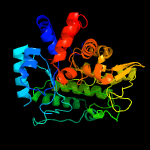
| 6 | c5jp0A_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase bogh3b;
PDBTitle: bacteroides ovatus xyloglucan pul gh3b with bound glucose
|
|
|
|
| 7 | c5vqdA_
|
|
 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucoside phosphorylase bglx;
PDBTitle: beta-glucoside phosphorylase bglx
|
|
|
|
| 8 | c5m6gA_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure glucan 1,4-beta-glucosidase from saccharopolyspora2 erythraea
|
|
|
|
| 9 | c5bu9B_
|
|
 |
100.0 |
28 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of beta-n-acetylhexosaminidase from beutenbergia2 cavernae dsm 12333
|
|
|
|
| 10 | c3f93D_
|
|
 |
100.0 |
21 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of exo-1,3/1,4-beta-glucanase (exop) from2 pseudoalteromonas sp. bb1
|
|
|
|
| 11 | c5z87B_
|
|
 |
100.0 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:emgh1;
PDBTitle: structural of a novel b-glucosidase emgh1 at 2.3 angstrom from2 erythrobacter marinus
|
|
|
|
| 12 | c5yotB_
|
|
 |
100.0 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:isoprimeverose-producing enzyme;
PDBTitle: isoprimeverose-producing enzyme from aspergillus oryzae in complex2 with isoprimeverose
|
|
|
|
| 13 | d1x38a1
|
|
 |
100.0 |
20 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:NagZ-like |
|
|
|
| 14 | c1ex1A_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:protein (beta-d-glucan exohydrolase isoenzyme exo1);
PDBTitle: beta-d-glucan exohydrolase from barley
|
|
|
|
| 15 | c3u48A_
|
|
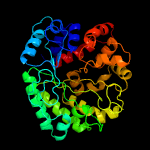 |
100.0 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:jmb19063;
PDBTitle: from soil to structure: a novel dimeric family 3-beta-glucosidase2 isolated from compost using metagenomic analysis
|
|
|
|
| 16 | c4zo9B_
|
|
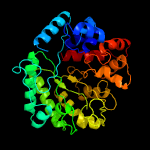 |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:lin1840 protein;
PDBTitle: crystal structure of mutant (d270a) beta-glucosidase from listeria2 innocua in complex with laminaribiose
|
|
|
|
| 17 | c5z9sB_
|
|
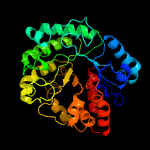 |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase family 3 protein;
PDBTitle: functional and structural characterization of a beta-glucosidase2 involved in saponin metabolism from intestinal bacteria
|
|
|
|
| 18 | c5tf0B_
|
|
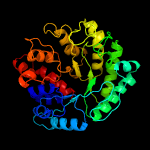 |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycosyl hydrolase family 3 n-terminal domain protein;
PDBTitle: crystal structure of glycosil hydrolase family 3 n-terminal domain2 protein from bacteroides intestinalis
|
|
|
|
| 19 | c3wo8B_
|
|
 |
100.0 |
31 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-n-acetylglucosaminidase;
PDBTitle: crystal structure of the beta-n-acetylglucosaminidase from thermotoga2 maritima
|
|
|
|
| 20 | c5bzaA_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-n-acetylhexosaminidase;
PDBTitle: crystal structure of cbsa from thermotoga neapolitana
|
|
|
|
| 21 | c5iobC_ |
|
not modelled |
100.0 |
41 |
PDB header:hydrolase
Chain: C: PDB Molecule:beta-glucosidase-related glycosidases;
PDBTitle: crystal structure of beta-n-acetylglucosaminidase-like protein from2 corynebacterium glutamicum
|
|
|
| 22 | c6q7jB_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:exo-1,4-beta-xylosidase xlnd;
PDBTitle: gh3 exo-beta-xylosidase (xlnd) in complex with xylobiose aziridine2 activity based probe
|
|
|
| 23 | c4iidB_ |
|
not modelled |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase 1;
PDBTitle: crystal structure of beta-glucosidase 1 from aspergillus aculeatus in2 complex with 1-deoxynojirimycin
|
|
|
| 24 | c5nbsA_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: structural studies of a glycoside hydrolase family 3 beta-glucosidase2 from the model fungus neurospora crassa
|
|
|
| 25 | c5a7mA_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-xylosidase;
PDBTitle: the structure of hypocrea jecorina beta-xylosidase xyl3a (bxl1)
|
|
|
| 26 | c3zz1A_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-d-glucoside glucohydrolase;
PDBTitle: crystal structure of a glycoside hydrolase family 3 beta-glucosidase,2 bgl1 from hypocrea jecorina at 2.1a resolution.
|
|
|
| 27 | c4d0jD_ |
|
not modelled |
100.0 |
22 |
PDB header:hydrolase
Chain: D: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of glycoside hydrolase family 3 beta-2 glucosidase cel3a from the moderately thermophilic fungus3 rasamsonia emersonii
|
|
|
| 28 | c4g6cA_ |
|
not modelled |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase 1;
PDBTitle: crystal structure of beta-hexosaminidase 1 from burkholderia2 cenocepacia j2315
|
|
|
| 29 | c2x41A_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: structure of beta-glucosidase 3b from thermotoga neapolitana in2 complex with glucose
|
|
|
| 30 | c3tevA_ |
|
not modelled |
100.0 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycosyl hyrolase, family 3;
PDBTitle: the crystal structure of glycosyl hydrolase from deinococcus2 radiodurans r1
|
|
|
| 31 | c5wabD_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: D: PDB Molecule:putative beta-glucosidase;
PDBTitle: crystal structure of bifidobacterium adolescentis gh3 beta-glucosidase
|
|
|
| 32 | c5wabC_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:putative beta-glucosidase;
PDBTitle: crystal structure of bifidobacterium adolescentis gh3 beta-glucosidase
|
|
|
| 33 | c4i3gB_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase;
PDBTitle: crystal structure of desr, a beta-glucosidase from streptomyces2 venezuelae in complex with d-glucose.
|
|
|
| 34 | c4gvgB_ |
|
not modelled |
100.0 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of salmonella typhimurium family 3 glycoside2 hydrolase (nagz)
|
|
|
| 35 | c3ac0B_ |
|
not modelled |
100.0 |
21 |
PDB header:hydrolase
Chain: B: PDB Molecule:beta-glucosidase i;
PDBTitle: crystal structure of beta-glucosidase from kluyveromyces marxianus in2 complex with glucose
|
|
|
| 36 | c5g1mA_ |
|
not modelled |
100.0 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-hexosaminidase;
PDBTitle: crystal structure of nagz from pseudomonas aeruginosa
|
|
|
| 37 | d1tr9a_ |
|
not modelled |
100.0 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:NagZ-like |
|
|
| 38 | c5wvpA_ |
|
not modelled |
100.0 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:beta-glucosidase;
PDBTitle: expression, characterization and crystal structure of a novel beta-2 glucosidase from paenibacillus barengoltzii
|
|
|
| 39 | c5k6lA_ |
|
not modelled |
100.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:b-glucosidase;
PDBTitle: structure of a gh3 b-glucosidase from cow rumen metagenome
|
|
|
| 40 | d1o4ua1 |
|
not modelled |
76.7 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:Nicotinate/Quinolinate PRTase C-terminal domain-like
Family:NadC C-terminal domain-like |
|
|
| 41 | c4ce4S_ |
|
not modelled |
65.4 |
22 |
PDB header:ribosome
Chain: S: PDB Molecule:mrpl18;
PDBTitle: 39s large subunit of the porcine mitochondrial ribosome
|
|
|
| 42 | d2p12a1 |
|
not modelled |
62.3 |
16 |
Fold:FomD barrel-like
Superfamily:FomD-like
Family:FomD-like |
|
|
| 43 | d2gycm1 |
|
not modelled |
62.0 |
20 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
|
|
| 44 | d1pv8a_ |
|
not modelled |
58.5 |
27 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase) |
|
|
| 45 | c2htmB_ |
|
not modelled |
54.2 |
21 |
PDB header:biosynthetic protein
Chain: B: PDB Molecule:thiazole biosynthesis protein thig;
PDBTitle: crystal structure of ttha0676 from thermus thermophilus hb8
|
|
|
| 46 | c1o4uA_ |
|
not modelled |
50.9 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:type ii quinolic acid phosphoribosyltransferase;
PDBTitle: crystal structure of a nicotinate nucleotide pyrophosphorylase2 (tm1645) from thermotoga maritima at 2.50 a resolution
|
|
|
| 47 | c4n6eA_ |
|
not modelled |
44.3 |
23 |
PDB header:lyase/biosynthetic protein
Chain: A: PDB Molecule:putative thiosugar synthase;
PDBTitle: crystal structure of amycolatopsis orientalis bexx/cyso complex
|
|
|
| 48 | d2vcha1 |
|
not modelled |
41.5 |
16 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
|
|
| 49 | c3bboQ_ |
|
not modelled |
41.4 |
17 |
PDB header:ribosome
Chain: Q: PDB Molecule:ribosomal protein l18;
PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
|
|
|
| 50 | c5nlmB_ |
|
not modelled |
41.4 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:indoxyl udp-glucosyltransferase;
PDBTitle: complex between a udp-glucosyltransferase from polygonum tinctorium2 capable of glucosylating indoxyl and indoxyl sulfate
|
|
|
| 51 | d1d2da_ |
|
not modelled |
39.1 |
20 |
Fold:S15/NS1 RNA-binding domain
Superfamily:S15/NS1 RNA-binding domain
Family:a tRNA synthase domain |
|
|
| 52 | d1xm3a_ |
|
not modelled |
37.9 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:ThiG-like
Family:ThiG-like |
|
|
| 53 | c2djvA_ |
|
not modelled |
37.2 |
17 |
PDB header:protein binding
Chain: A: PDB Molecule:methionyl-trna synthetase;
PDBTitle: solution structures of the whep-trs domain of human2 methionyl-trna synthetase
|
|
|
| 54 | d1fyja_ |
|
not modelled |
36.7 |
27 |
Fold:S15/NS1 RNA-binding domain
Superfamily:S15/NS1 RNA-binding domain
Family:a tRNA synthase domain |
|
|
| 55 | c4v19S_ |
|
not modelled |
34.5 |
18 |
PDB header:ribosome
Chain: S: PDB Molecule:mitoribosomal protein ul18m, mrpl18;
PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
|
|
|
| 56 | c3wdjA_ |
|
not modelled |
33.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:type i pullulanase;
PDBTitle: crystal structure of pullulanase complexed with maltotetraose from2 anoxybacillus sp. lm18-11
|
|
|
| 57 | c2jtxA_ |
|
not modelled |
31.9 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:transcription initiation factor iie subunit
PDBTitle: nmr structure of the tfiie-alpha carboxyl terminus
|
|
|
| 58 | c2rnqA_ |
|
not modelled |
29.7 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:transcription initiation factor iie subunit
PDBTitle: solution structure of the c-terminal acidic domain of tfiie2 alpha
|
|
|
| 59 | c1x59A_ |
|
not modelled |
29.3 |
11 |
PDB header:protein binding
Chain: A: PDB Molecule:histidyl-trna synthetase;
PDBTitle: solution structures of the whep-trs domain of human2 histidyl-trna synthetase
|
|
|
| 60 | c6jtdB_ |
|
not modelled |
28.3 |
19 |
PDB header:transferase
Chain: B: PDB Molecule:c-glycosyltransferase;
PDBTitle: crystal structure of tccgt1 in complex with udp
|
|
|
| 61 | d2c1xa1 |
|
not modelled |
27.0 |
7 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
|
|
| 62 | c4a3uB_ |
|
not modelled |
26.5 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadh\:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of the old yellow enzyme homologue from zymomonas mobilis2 (ncr)
|
|
|
| 63 | c2cosA_ |
|
not modelled |
25.4 |
26 |
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine protein kinase lats2;
PDBTitle: solution structure of rsgi ruh-038, a uba domain from mouse2 lats2 (large tumor suppressor homolog 2)
|
|
|
| 64 | d1r6ta1 |
|
not modelled |
24.8 |
23 |
Fold:S15/NS1 RNA-binding domain
Superfamily:S15/NS1 RNA-binding domain
Family:a tRNA synthase domain |
|
|
| 65 | d1yxya1 |
|
not modelled |
24.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:NanE-like |
|
|
| 66 | d1vhna_ |
|
not modelled |
24.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 67 | c2e8yA_ |
|
not modelled |
23.1 |
29 |
PDB header:hydrolase
Chain: A: PDB Molecule:amyx protein;
PDBTitle: crystal structure of pullulanase type i from bacillus subtilis str.2 168
|
|
|
| 68 | c5v2kA_ |
|
not modelled |
22.9 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:udp-glycosyltransferase 74f2;
PDBTitle: crystal structure of udp-glucosyltransferase, ugt74f2 (t15a), with udp2 and 2-bromobenzoic acid
|
|
|
| 69 | d1ovya_ |
|
not modelled |
22.7 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Translational machinery components
Family:Ribosomal protein L18 and S11 |
|
|
| 70 | c1w1nA_ |
|
not modelled |
22.2 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidylinositol 3-kinase tor1;
PDBTitle: the solution structure of the fatc domain of the protein kinase tor12 from yeast
|
|
|
| 71 | c2ya0A_ |
|
not modelled |
21.3 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative alkaline amylopullulanase;
PDBTitle: catalytic module of the multi-modular glycogen-degrading pneumococcal2 virulence factor spua
|
|
|
| 72 | d1k3ka_ |
|
not modelled |
21.0 |
33 |
Fold:Toxins' membrane translocation domains
Superfamily:Bcl-2 inhibitors of programmed cell death
Family:Bcl-2 inhibitors of programmed cell death |
|
|
| 73 | c3wc4A_ |
|
not modelled |
20.7 |
5 |
PDB header:transferase
Chain: A: PDB Molecule:udp-glucose:anthocyanidin 3-o-glucosyltransferase;
PDBTitle: crystal structure of udp-glucose: anthocyanidin 3-o-2 glucosyltransferase from clitoria ternatea
|
|
|
| 74 | c3j3vO_ |
|
not modelled |
20.3 |
18 |
PDB header:ribosome
Chain: O: PDB Molecule:50s ribosomal protein l18;
PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
|
|
|
| 75 | d2odfa1 |
|
not modelled |
19.8 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Zn-dependent exopeptidases
Family:FGase-like |
|
|
| 76 | c2y69X_ |
|
not modelled |
19.7 |
22 |
PDB header:electron transport
Chain: X: PDB Molecule:cytochrome c oxidase polypeptide 7b;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular oxygen
|
|
|
| 77 | d1v54k_ |
|
not modelled |
19.7 |
22 |
Fold:Single transmembrane helix
Superfamily:Mitochondrial cytochrome c oxidase subunit VIIb
Family:Mitochondrial cytochrome c oxidase subunit VIIb |
|
|
| 78 | c2kitA_ |
|
not modelled |
19.2 |
7 |
PDB header:transferase
Chain: A: PDB Molecule:serine/threonine-protein kinase tor1;
PDBTitle: the solution structure of the reduced yeast tor1 fatc domain bound to2 dpc micelles at 298k
|
|
|
| 79 | d2acva1 |
|
not modelled |
18.5 |
14 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
|
|
| 80 | c1iq8B_ |
|
not modelled |
18.1 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:archaeosine trna-guanine transglycosylase;
PDBTitle: crystal structure of archaeosine trna-guanine transglycosylase from2 pyrococcus horikoshii
|
|
|
| 81 | c4j7rA_ |
|
not modelled |
18.1 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:isoamylase;
PDBTitle: crystal structure of chlamydomonas reinhardtii isoamylase 1 (isa1)
|
|
|
| 82 | c5emiA_ |
|
not modelled |
17.6 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:cell wall hydrolase/autolysin;
PDBTitle: n-acetylmuramoyl-l-alanine amidase amic2 of nostoc punctiforme
|
|
|
| 83 | d2pq6a1 |
|
not modelled |
17.5 |
11 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:UDPGT-like |
|
|
| 84 | c2wanA_ |
|
not modelled |
17.2 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:pullulanase;
PDBTitle: pullulanase from bacillus acidopullulyticus
|
|
|
| 85 | c4jicB_ |
|
not modelled |
16.7 |
11 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gtn reductase;
PDBTitle: glycerol trinitrate reductase nera from agrobacterium radiobacter
|
|
|
| 86 | c3hbjA_ |
|
not modelled |
16.5 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:flavonoid 3-o-glucosyltransferase;
PDBTitle: structure of ugt78g1 complexed with udp
|
|
|
| 87 | c3atyA_ |
|
not modelled |
16.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:prostaglandin f2a synthase;
PDBTitle: crystal structure of tcoye
|
|
|
| 88 | c3w1hB_ |
|
not modelled |
16.1 |
17 |
PDB header:transferase
Chain: B: PDB Molecule:l-seryl-trna(sec) selenium transferase;
PDBTitle: crystal structure of the selenocysteine synthase sela from aquifex2 aeolicus
|
|
|
| 89 | c4binA_ |
|
not modelled |
15.9 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:n-acetylmuramoyl-l-alanine amidase amic;
PDBTitle: crystal structure of the e. coli n-acetylmuramoyl-l-alanine amidase2 amic
|
|
|
| 90 | c6ei9A_ |
|
not modelled |
15.9 |
15 |
PDB header:flavoprotein
Chain: A: PDB Molecule:trna-dihydrouridine synthase b;
PDBTitle: crystal structure of e. coli trna-dihydrouridine synthase b (dusb)
|
|
|
| 91 | c2qlcC_ |
|
not modelled |
15.5 |
13 |
PDB header:dna binding protein
Chain: C: PDB Molecule:dna repair protein radc homolog;
PDBTitle: the crystal structure of dna repair protein radc from chlorobium2 tepidum tls
|
|
|
| 92 | c5tmdA_ |
|
not modelled |
14.5 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:glycosyltransferase, os79;
PDBTitle: crystal structure of os79 from o. sativa in complex with u2f and2 trichothecene.
|
|
|
| 93 | c2h90A_ |
|
not modelled |
14.5 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xenobiotic reductase a;
PDBTitle: xenobiotic reductase a in complex with coumarin
|
|
|
| 94 | d1ulza2 |
|
not modelled |
14.4 |
18 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:BC N-terminal domain-like |
|
|
| 95 | c3ht4B_ |
|
not modelled |
14.1 |
23 |
PDB header:lyase
Chain: B: PDB Molecule:aluminum resistance protein;
PDBTitle: crystal structure of the q81a77_baccr protein from bacillus cereus.2 northeast structural genomics consortium target bcr213
|
|
|
| 96 | c5o60P_ |
|
not modelled |
14.1 |
16 |
PDB header:ribosome
Chain: P: PDB Molecule:50s ribosomal protein l18;
PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
|
|
|
| 97 | d1gwja_ |
|
not modelled |
13.9 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:FMN-linked oxidoreductases
Family:FMN-linked oxidoreductases |
|
|
| 98 | d1n82a_ |
|
not modelled |
13.9 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:beta-glycanases |
|
|
| 99 | c5hssA_ |
|
not modelled |
13.9 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:linalool dehydratase/isomerase;
PDBTitle: linalool dehydratase/isomerase: ldi with monoterpene substrate
|
|
|