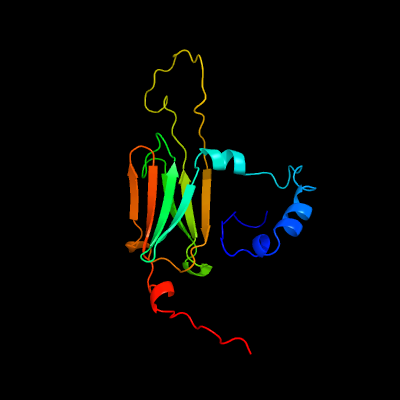
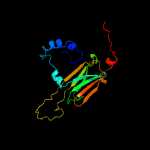
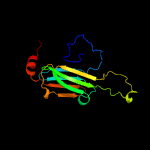
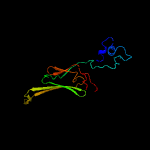
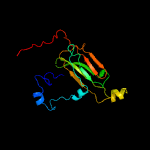
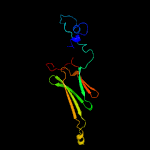
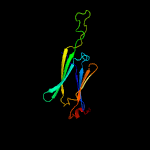
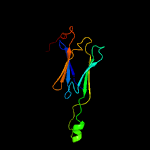
1 d1gmea_
99.9
23
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: HSP20
2 c3w1zA_
99.9
28
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 16;PDBTitle: heat shock protein 16.0 from schizosaccharomyces pombe
3 c2ygdV_
99.9
26
PDB header: chaperoneChain: V: PDB Molecule: PDBTitle: molecular architectures of the 24meric eye lens chaperone alphab-2 crystallin elucidated by a triple hybrid approach
4 c5mb8J_
99.9
19
PDB header: chaperoneChain: J: PDB Molecule: 25.3 kda heat shock protein, chloroplastic;PDBTitle: hsp21 dodecamer, structural model based on cryo-em and homology2 modelling
5 c6dv5T_
99.9
22
PDB header: chaperoneChain: T: PDB Molecule: heat shock protein beta-1;PDBTitle: oligomeric complex of a hsp27 24-mer at 3.6 a resolution
6 c3w1zD_
99.9
31
PDB header: chaperoneChain: D: PDB Molecule: heat shock protein 16;PDBTitle: heat shock protein 16.0 from schizosaccharomyces pombe
7 d1gmeb_
99.9
30
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: HSP20
8 d1shsa_
99.9
23
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: HSP20
9 c1shsD_
99.9
23
PDB header: heat shock proteinChain: D: PDB Molecule: small heat shock protein;PDBTitle: small heat shock protein from methanococcus jannaschii
10 c3glaA_
99.9
33
PDB header: chaperoneChain: A: PDB Molecule: low molecular weight heat shock protein;PDBTitle: crystal structure of the hspa from xanthomonas axonopodis
11 c3l1eA_
99.9
24
PDB header: chaperoneChain: A: PDB Molecule: alpha-crystallin a chain;PDBTitle: bovine alphaa crystallin zinc bound
12 c4feiA_
99.9
31
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein-related protein;PDBTitle: hsp17.7 from deinococcus radiodurans
13 c4ydzA_
99.9
23
PDB header: chaperoneChain: A: PDB Molecule: stress-induced protein 1;PDBTitle: stress-induced protein 1 from caenorhabditis elegans
14 c5mb8L_
99.8
25
PDB header: chaperoneChain: L: PDB Molecule: 25.3 kda heat shock protein, chloroplastic;PDBTitle: hsp21 dodecamer, structural model based on cryo-em and homology2 modelling
15 c2bolA_
99.8
17
PDB header: heat shock proteinChain: A: PDB Molecule: small heat shock protein;PDBTitle: crystal structure and assembly of tsp36, a metazoan small heat shock2 protein
16 c6ewnA_
99.8
32
PDB header: chaperoneChain: A: PDB Molecule: hspa;PDBTitle: hspa from thermosynechococcus vulcanus in the presence of 2m urea with2 initial stages of denaturation
17 d2h50a1
99.8
32
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: HSP20
18 c5ds1B_
99.8
30
PDB header: chaperoneChain: B: PDB Molecule: 17.1 kda class ii heat shock protein;PDBTitle: core domain of the class ii small heat-shock protein hsp 17.7 from2 pisum sativum
19 c4zjdF_
99.8
31
PDB header: chaperoneChain: F: PDB Molecule: aggregation suppressing protein;PDBTitle: small heat shock protein agsa from salmonella typhimurium: truncations2 at n- and c- termini
20 c3aabA_
99.8
25
PDB header: chaperoneChain: A: PDB Molecule: putative uncharacterized protein st1653;PDBTitle: small heat shock protein hsp14.0 with the mutations of i120f and i122f2 in the form i crystal
21 c5ltwK_
not modelled
99.8
21
PDB header: protein bindingChain: K: PDB Molecule: heat shock protein beta-6;PDBTitle: complex of human 14-3-3 sigma clu1 mutant with phosphorylated heat2 shock protein b6
22 c4ylcF_
not modelled
99.8
25
PDB header: chaperoneChain: F: PDB Molecule: heat shock protein hsp20;PDBTitle: crystal structure of del-c4 mutant of hsp14.1 from sulfolobus2 solfatataricus p2
23 c6f2rK_
not modelled
99.8
17
PDB header: chaperoneChain: K: PDB Molecule: hspb2,heat shock protein beta-2,heat shock protein beta-2,PDBTitle: a hetrotetramer of human hspb2 and hspb3
24 c6f2rE_
not modelled
99.8
19
PDB header: chaperoneChain: E: PDB Molecule: hspb2,heat shock protein beta-2,heat shock protein beta-2,PDBTitle: a hetrotetramer of human hspb2 and hspb3
25 c2wj5A_
not modelled
99.8
22
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein beta-6;PDBTitle: rat alpha crystallin domain
26 c6f2rI_
not modelled
99.8
16
PDB header: chaperoneChain: I: PDB Molecule: hspb2,heat shock protein beta-2,heat shock protein beta-2,PDBTitle: a hetrotetramer of human hspb2 and hspb3
27 c4rzkA_
not modelled
99.8
25
PDB header: chaperoneChain: A: PDB Molecule: small heat shock protein hsp20 family;PDBTitle: crystal structure of sulfolobus solfataricus hsp20.1 acd
28 c2n3jA_
not modelled
99.8
24
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein beta-1;PDBTitle: solution structure of the alpha-crystallin domain from the redox-2 sensitive chaperone, hspb1
29 c5zulB_
not modelled
99.8
26
PDB header: chaperoneChain: B: PDB Molecule: small heat shock protein;PDBTitle: small heat shock protein from mycobacterium marinum m : form-3
30 c6f2rA_
not modelled
99.7
19
PDB header: chaperoneChain: A: PDB Molecule: hspb2,heat shock protein beta-2,heat shock protein beta-2,PDBTitle: a hetrotetramer of human hspb2 and hspb3
31 c2klrA_
not modelled
99.7
28
PDB header: chaperoneChain: A: PDB Molecule: alpha-crystallin b chain;PDBTitle: solid-state nmr structure of the alpha-crystallin domain in alphab-2 crystallin oligomers
32 c3q9qB_
not modelled
99.7
25
PDB header: chaperoneChain: B: PDB Molecule: heat shock protein beta-1;PDBTitle: hspb1 fragment second crystal form
33 c6f2rT_
not modelled
99.6
22
PDB header: chaperoneChain: T: PDB Molecule: heat shock protein beta-3,heat shock protein beta-3,heatPDBTitle: a hetrotetramer of human hspb2 and hspb3
34 c2wj7D_
not modelled
99.5
30
PDB header: chaperoneChain: D: PDB Molecule: alpha-crystallin b chain;PDBTitle: human alphab crystallin
35 c4pbdA_
not modelled
99.4
23
PDB header: protein bindingChain: A: PDB Molecule: protein shq1 homolog;PDBTitle: crystal structure of the n-terminal cs domain of human shq1
36 c6gxzD_
not modelled
99.1
22
PDB header: chaperoneChain: D: PDB Molecule: pih1 domain-containing protein 1;PDBTitle: crystal structure of the human rpap3(tpr2)-pih1d1(cs) complex
37 d1rl1a_
not modelled
98.0
16
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: GS domain
38 c2jkiS_
not modelled
97.9
17
PDB header: chaperoneChain: S: PDB Molecule: sgt1-like protein;PDBTitle: complex of hsp90 n-terminal and sgt1 cs domain
39 c3eudE_
not modelled
97.3
18
PDB header: nuclear proteinChain: E: PDB Molecule: protein shq1;PDBTitle: structure of the cs domain of the essential h/aca rnp assembly protein2 shq1p
40 c3igfB_
not modelled
97.2
31
PDB header: atp binding proteinChain: B: PDB Molecule: all4481 protein;PDBTitle: crystal structure of the all4481 protein from nostoc sp. pcc 7120,2 northeast structural genomics consortium target nsr300
41 d1ejfa_
not modelled
96.6
12
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: Co-chaperone p23-like
42 c1x5mA_
not modelled
96.5
8
PDB header: apoptosis, signaling proteinChain: A: PDB Molecule: calcyclin-binding protein;PDBTitle: solution structure of the core domain of calcyclin binding2 protein; siah-interacting protein (sip)
43 c6mv2A_
not modelled
96.2
8
PDB header: oxidoreductaseChain: A: PDB Molecule: cytochrome b5 reductase 4;PDBTitle: 2.05a resolution structure of the cs-b5r domains of human ncb5or2 (nadp+ form)
44 c2k8qA_
not modelled
96.2
17
PDB header: structural proteinChain: A: PDB Molecule: protein shq1;PDBTitle: nmr structure of shq1p n-terminal domain
45 c2o30A_
not modelled
96.2
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: nuclear movement protein;PDBTitle: nuclear movement protein from e. cuniculi gb-m1
46 d1wgva_
not modelled
96.1
17
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: Nuclear movement domain
47 d1wfia_
not modelled
95.9
21
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: Nuclear movement domain
48 d1xo9a_
not modelled
95.1
20
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: Co-chaperone p23-like
49 d1wh0a_
not modelled
94.6
16
Fold: HSP20-like chaperonesSuperfamily: HSP20-like chaperonesFamily: GS domain
50 c2rh0B_
not modelled
82.6
13
PDB header: nuclear proteinChain: B: PDB Molecule: nudc domain-containing protein 2;PDBTitle: crystal structure of nudc domain-containing protein 2 (13542905) from2 mus musculus at 1.95 a resolution
51 c2cg9Y_
not modelled
62.4
10
PDB header: chaperoneChain: Y: PDB Molecule: co-chaperone protein sba1;PDBTitle: crystal structure of an hsp90-sba1 closed chaperone complex
52 d2fqla1
not modelled
61.2
17
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like
53 d1vyfa_
not modelled
46.2
14
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
54 d2ga5a1
not modelled
43.0
20
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like
55 d1ekga_
not modelled
41.9
41
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like
56 d1eala_
not modelled
40.8
17
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
57 c4jpdA_
not modelled
40.4
24
PDB header: metal binding proteinChain: A: PDB Molecule: protein cyay;PDBTitle: the structure of cyay from burkholderia cenocepacia
58 c4i3dB_
not modelled
40.0
14
PDB header: fluorescent proteinChain: B: PDB Molecule: bilirubin-inducible fluorescent protein unag;PDBTitle: crystal structure of fluorescent protein unag n57a mutant
59 c3em0A_
not modelled
39.3
19
PDB header: lipid binding proteinChain: A: PDB Molecule: ileal bile acid-binding protein;PDBTitle: crystal structure of zebrafish ileal bile acid-bindin protein2 complexed with cholic acid (crystal form b).
60 d1o1va_
not modelled
39.3
15
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
61 c4hs5B_
not modelled
39.2
13
PDB header: metal binding proteinChain: B: PDB Molecule: protein cyay;PDBTitle: frataxin from psychromonas ingrahamii as a model to study stability2 modulation within cyay protein family
62 c6fcoB_
not modelled
36.0
28
PDB header: transport proteinChain: B: PDB Molecule: mitochondrial frataxin-like protein;PDBTitle: structural and functional characterisation of frataxin (fxn) like2 protein from chaetomium thermophilum
63 d2a0aa1
not modelled
35.2
15
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
64 c6c1zA_
not modelled
34.4
13
PDB header: lipid binding proteinChain: A: PDB Molecule: lipid binding protein;PDBTitle: crystal structure of apo caenorhabditis elegans lipid binding protein2 8 (lbp-8)
65 d2j01h1
not modelled
34.1
9
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L6
66 d2ftba1
not modelled
33.8
29
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
67 c5bvtA_
not modelled
33.4
15
PDB header: lipid binding proteinChain: A: PDB Molecule: epidermal fatty acid-binding protein;PDBTitle: palmitate-bound pfabp5
68 d1ggla_
not modelled
32.0
20
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
69 c2lbaA_
not modelled
31.7
14
PDB header: lipid binding proteinChain: A: PDB Molecule: babp protein;PDBTitle: solution structure of chicken ileal babp in complex with2 glycochenodeoxycholic acid
70 d1ew4a_
not modelled
31.3
33
Fold: N domain of copper amine oxidase-likeSuperfamily: Frataxin/Nqo15-likeFamily: Frataxin-like
71 d1yiva1
not modelled
29.2
21
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
72 d1tw4a_
not modelled
29.1
31
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
73 d2cqla1
not modelled
28.8
11
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L6
74 d1p6pa_
not modelled
28.8
26
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
75 c5kz5H_
not modelled
28.7
41
PDB header: transferase/oxidoreductaseChain: H: PDB Molecule: frataxin, mitochondrial;PDBTitle: architecture of the human mitochondrial iron-sulfur cluster assembly2 machinery: the complex formed by the iron donor, the sulfur donor,3 and the scaffold
76 d1crba_
not modelled
28.4
24
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
77 d2f73a1
not modelled
28.3
14
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
78 d1bwya_
not modelled
27.9
17
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
79 c5xnaB_
not modelled
26.7
11
PDB header: lipid transportChain: B: PDB Molecule: sahs1;PDBTitle: crystal structure of a secretary abundant heat soluble (sahs) protein2 from ramazzottius varieornatus (from dimer sample)
80 c2q9sA_
not modelled
26.5
26
PDB header: lipid binding proteinChain: A: PDB Molecule: fatty acid-binding protein;PDBTitle: linoleic acid bound to fatty acid binding protein 4
81 d1pmpa_
not modelled
26.0
21
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
82 d1mdca_
not modelled
25.9
17
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
83 d1rl6a1
not modelled
25.9
9
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L6
84 d1lfoa_
not modelled
24.8
14
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
85 c4nkbB_
not modelled
24.3
31
PDB header: transferaseChain: B: PDB Molecule: probable serine/threonine-protein kinase zyg-1;PDBTitle: crystal structure of the cryptic polo box (cpb)of zyg-1
86 d2hnxa1
not modelled
24.1
24
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
87 d1kqwa_
not modelled
24.1
17
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
88 d1kzwa_
not modelled
23.8
14
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
89 d1lpja_
not modelled
23.6
14
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
90 d1fdqa_
not modelled
23.5
19
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
91 d1b56a_
not modelled
23.0
14
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
92 d1qw9a1
not modelled
22.5
10
Fold: Glycosyl hydrolase domainSuperfamily: Glycosyl hydrolase domainFamily: Composite domain of glycosyl hydrolase families 5, 30, 39 and 51
93 d1vqoe1
not modelled
21.8
26
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L6
94 c5fu4B_
not modelled
21.2
34
PDB header: sugar binding proteinChain: B: PDB Molecule: cbm74-rfgh5;PDBTitle: the complexity of the ruminococcus flavefaciens cellulosome reflects2 an expansion in glycan recognition
95 d1g7na_
not modelled
19.5
26
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
96 d2zjre2
not modelled
17.2
9
Fold: Ribosomal protein L6Superfamily: Ribosomal protein L6Family: Ribosomal protein L6
97 d1ifca_
not modelled
15.4
15
Fold: LipocalinsSuperfamily: LipocalinsFamily: Fatty acid binding protein-like
98 c3pcsB_
not modelled
15.2
15
PDB header: protein transport/transferaseChain: B: PDB Molecule: espg;PDBTitle: structure of espg-pak2 autoinhibitory ialpha3 helix complex
99 c2n93A_
not modelled
14.9
11
PDB header: lipid binding proteinChain: A: PDB Molecule: fatty acid-binding protein;PDBTitle: solution structure of lcfabp