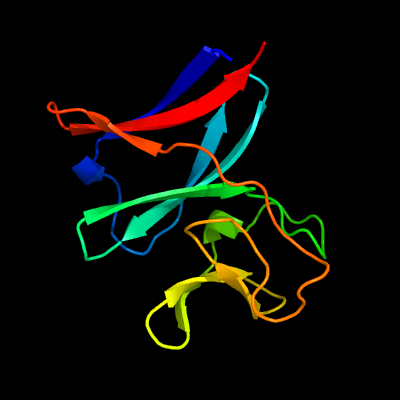
| 1 | c4aivA_
|
|
 |
100.0 |
100 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:probable nitrite reductase [nad(p)h] small subunit nird;
PDBTitle: crystal structure of putative nadh-dependent nitrite reductase small2 subunit from mycobacterium tuberculosis
|
|
|
|
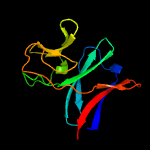
| 2 | d2jo6a1
|
|
 |
100.0 |
41 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
|
|
|
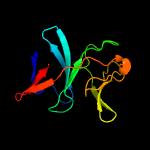
| 3 | d3c0da1
|
|
 |
100.0 |
38 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
|
|
|
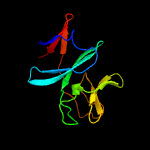
| 4 | d2jzaa1
|
|
 |
100.0 |
44 |
Fold:ISP domain
Superfamily:ISP domain
Family:NirD-like |
|
|
|
| 5 | d1vm9a_
|
|
 |
100.0 |
18 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
|
| 6 | c3gceA_
|
|
 |
99.9 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ferredoxin component of carbazole 1,9a-
PDBTitle: ferredoxin of carbazole 1,9a-dioxygenase from nocardioides2 aromaticivorans ic177
|
|
|
|
| 7 | c2de7E_
|
|
 |
99.9 |
24 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ferredoxin component of carbazole;
PDBTitle: the substrate-bound complex between oxygenase and2 ferredoxin in carbazole 1,9a-dioxygenase
|
|
|
|
| 8 | c4qdfA_
|
|
 |
99.9 |
17 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: A: PDB Molecule:3-ketosteroid 9alpha-hydroxylase oxygenase;
PDBTitle: crystal structure of apo ksha5 and ksha1 in complex with 1,4-30q-coa2 from r. rhodochrous
|
|
|
|
| 9 | c4qdfB_
|
|
 |
99.9 |
15 |
PDB header:oxidoreductase/oxidoreductase inhibitor
Chain: B: PDB Molecule:3-ketosteroid 9alpha-hydroxylase oxygenase;
PDBTitle: crystal structure of apo ksha5 and ksha1 in complex with 1,4-30q-coa2 from r. rhodochrous
|
|
|
|
| 10 | c2qpzA_
|
|
 |
99.9 |
24 |
PDB header:metal binding protein
Chain: A: PDB Molecule:naphthalene 1,2-dioxygenase system ferredoxin
PDBTitle: naphthalene 1,2-dioxygenase rieske ferredoxin
|
|
|
|
| 11 | d1fqta_
|
|
 |
99.9 |
19 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
|
| 12 | c2zylA_
|
|
 |
99.9 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:possible oxidoreductase;
PDBTitle: crystal structure of 3-ketosteroid-9-alpha-hydroxylase2 (ksha) from m. tuberculosis
|
|
|
|
| 13 | c2de7B_
|
|
 |
99.9 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:terminal oxygenase component of carbazole;
PDBTitle: the substrate-bound complex between oxygenase and2 ferredoxin in carbazole 1,9a-dioxygenase
|
|
|
|
| 14 | c3gkqB_
|
|
 |
99.9 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:terminal oxygenase component of carbazole 1,9a-dioxygenase;
PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from novosphingobium2 sp. ka1
|
|
|
|
| 15 | c3d89A_
|
|
 |
99.9 |
20 |
PDB header:electron transport
Chain: A: PDB Molecule:rieske domain-containing protein;
PDBTitle: crystal structure of a soluble rieske ferredoxin from mus musculus
|
|
|
|
| 16 | d2de6a1
|
|
 |
99.9 |
15 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
|
|
|
| 17 | d1z01a1
|
|
 |
99.9 |
20 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
|
|
|
| 18 | c3gcfC_
|
|
 |
99.9 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:terminal oxygenase component of carbazole 1,9a-
PDBTitle: terminal oxygenase of carbazole 1,9a-dioxygenase from2 nocardioides aromaticivorans ic177
|
|
|
|
| 19 | c3n0qA_
|
|
 |
99.9 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative aromatic-ring hydroxylating dioxygenase;
PDBTitle: crystal structure of a putative aromatic-ring hydroxylating2 dioxygenase (tm1040_3219) from silicibacter sp. tm1040 at 1.80 a3 resolution
|
|
|
|
| 20 | c3vcaA_
|
|
 |
99.9 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ring-hydroxylating dioxygenase;
PDBTitle: quaternary ammonium oxidative demethylation: x-ray crystallographic,2 resonance raman and uv-visible spectroscopic analysis of a rieske-3 type demethylase
|
|
|
|
| 21 | c1z01D_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:2-oxo-1,2-dihydroquinoline 8-monooxygenase, oxygenase
PDBTitle: 2-oxoquinoline 8-monooxygenase component: active site modulation by2 rieske-[2fe-2s] center oxidation/reduction
|
|
|
| 22 | c3gteB_ |
|
not modelled |
99.9 |
27 |
PDB header:electron transport, oxidoreductase
Chain: B: PDB Molecule:ddmc;
PDBTitle: crystal structure of dicamba monooxygenase with non-heme iron
|
|
|
| 23 | c2i7fB_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:ferredoxin component of dioxygenase;
PDBTitle: sphingomonas yanoikuyae b1 ferredoxin
|
|
|
| 24 | c3dqyA_ |
|
not modelled |
99.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:toluene 1,2-dioxygenase system ferredoxin
PDBTitle: crystal structure of toluene 2,3-dioxygenase ferredoxin
|
|
|
| 25 | d1ulia1 |
|
not modelled |
99.9 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
|
|
| 26 | d1wqla1 |
|
not modelled |
99.9 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
|
|
| 27 | c5cxmC_ |
|
not modelled |
99.9 |
15 |
PDB header:metal binding protein
Chain: C: PDB Molecule:cytochrome b6/f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cyanobacterial plasma membrane rieske protein2 petc3 from synechocystis pcc 6803
|
|
|
| 28 | d2bmoa1 |
|
not modelled |
99.9 |
17 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
|
|
| 29 | c1uljA_ |
|
not modelled |
99.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:biphenyl dioxygenase large subunit;
PDBTitle: biphenyl dioxygenase (bpha1a2) in complex with the substrate
|
|
|
| 30 | c1wqlA_ |
|
not modelled |
99.9 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:iron-sulfur protein large subunit of cumene dioxygenase;
PDBTitle: cumene dioxygenase (cuma1a2) from pseudomonas fluorescens ip01
|
|
|
| 31 | d2b1xa1 |
|
not modelled |
99.9 |
17 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
|
|
| 32 | d1o7na1 |
|
not modelled |
99.9 |
17 |
Fold:ISP domain
Superfamily:ISP domain
Family:Ring hydroxylating alpha subunit ISP domain |
|
|
| 33 | c2gbxE_ |
|
not modelled |
99.9 |
20 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:biphenyl 2,3-dioxygenase alpha subunit;
PDBTitle: crystal structure of biphenyl 2,3-dioxygenase from sphingomonas2 yanoikuyae b1 bound to biphenyl
|
|
|
| 34 | c2hmnA_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:naphthalene 1,2-dioxygenase alpha subunit;
PDBTitle: crystal structure of the naphthalene 1,2-dioxygenase f352v2 mutant bound to anthracene.
|
|
|
| 35 | c2b1xE_ |
|
not modelled |
99.9 |
17 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:naphthalene dioxygenase large subunit;
PDBTitle: crystal structure of naphthalene 1,2-dioxygenase from rhodococcus sp.
|
|
|
| 36 | c4aayH_ |
|
not modelled |
99.9 |
19 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:arob;
PDBTitle: crystal structure of the arsenite oxidase protein complex2 from rhizobium species strain nt-26
|
|
|
| 37 | d1rfsa_ |
|
not modelled |
99.8 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 38 | d1g8kb_ |
|
not modelled |
99.8 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 39 | d1q90c_ |
|
not modelled |
99.8 |
13 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 40 | d3cx5e1 |
|
not modelled |
99.8 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 41 | d2e74d1 |
|
not modelled |
99.8 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 42 | d1riea_ |
|
not modelled |
99.7 |
14 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 43 | d1nyka_ |
|
not modelled |
99.7 |
21 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 44 | c2e76D_ |
|
not modelled |
99.6 |
14 |
PDB header:photosynthesis
Chain: D: PDB Molecule:cytochrome b6-f complex iron-sulfur subunit;
PDBTitle: crystal structure of the cytochrome b6f complex with tridecyl-2 stigmatellin (tds) from m.laminosus
|
|
|
| 45 | c2fyuE_ |
|
not modelled |
99.6 |
13 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit,
PDBTitle: crystal structure of bovine heart mitochondrial bc1 with jg1442 inhibitor
|
|
|
| 46 | c1p84E_ |
|
not modelled |
99.6 |
16 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
|
|
|
| 47 | c2fynO_ |
|
not modelled |
99.5 |
19 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur
PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
|
|
|
| 48 | c2nvgA_ |
|
not modelled |
99.5 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: soluble domain of rieske iron sulfur protein.
|
|
|
| 49 | d1jm1a_ |
|
not modelled |
99.4 |
22 |
Fold:ISP domain
Superfamily:ISP domain
Family:Rieske iron-sulfur protein (ISP) |
|
|
| 50 | c6hwhB_ |
|
not modelled |
99.4 |
13 |
PDB header:electron transport
Chain: B: PDB Molecule:ubiquinol-cytochrome c reductase iron-sulfur subunit;
PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
|
|
|
| 51 | c6fo2R_ |
|
not modelled |
98.9 |
16 |
PDB header:membrane protein
Chain: R: PDB Molecule:cytochrome b-c1 complex subunit rieske, mitochondrial;
PDBTitle: cryoem structure of bovine cytochrome bc1 with no ligand bound
|
|
|
| 52 | d2hf1a1 |
|
not modelled |
57.0 |
19 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
|
|
| 53 | d2jnya1 |
|
not modelled |
56.9 |
19 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
|
|
| 54 | d2pk7a1 |
|
not modelled |
53.6 |
16 |
Fold:Trm112p-like
Superfamily:Trm112p-like
Family:Trm112p-like |
|
|
| 55 | c2jr6A_ |
|
not modelled |
48.5 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0434 protein nma0874;
PDBTitle: solution structure of upf0434 protein nma0874. northeast structural2 genomics target mr32
|
|
|
| 56 | c2kpiA_ |
|
not modelled |
46.9 |
19 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein sco3027;
PDBTitle: solution nmr structure of streptomyces coelicolor sco30272 modeled with zn+2 bound, northeast structural genomics3 consortium target rr58
|
|
|
| 57 | c2js4A_ |
|
not modelled |
46.0 |
22 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:upf0434 protein bb2007;
PDBTitle: solution nmr structure of bordetella bronchiseptica protein2 bb2007. northeast structural genomics consortium target3 bor54
|
|
|
| 58 | c6f5zC_ |
|
not modelled |
40.0 |
13 |
PDB header:transferase
Chain: C: PDB Molecule:upf0434 family protein;
PDBTitle: complex between the haloferax volcanii trm112 methyltransferase2 activator and the hvo_0019 putative methyltransferase
|
|
|
| 59 | c3tenD_ |
|
not modelled |
33.5 |
0 |
PDB header:hydrolase
Chain: D: PDB Molecule:cs2 hydrolase;
PDBTitle: holo form of carbon disulfide hydrolase
|
|
|
| 60 | c4esjA_ |
|
not modelled |
31.0 |
19 |
PDB header:hydrolase/dna
Chain: A: PDB Molecule:type-2 restriction enzyme dpni;
PDBTitle: restriction endonuclease dpni in complex with target dna
|
|
|
| 61 | c3wwnB_ |
|
not modelled |
27.3 |
13 |
PDB header:metal binding protein/transferase
Chain: B: PDB Molecule:orff;
PDBTitle: crystal structure of lysz from thermus thermophilus complex with lysw
|
|
|
| 62 | c3vrkA_ |
|
not modelled |
25.4 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:carbonyl sulfide hydrolase;
PDBTitle: crystal structutre of thiobacillus thioparus thi115 carbonyl sulfide2 hydrolase / thiocyanate complex
|
|
|
| 63 | c3vpbF_ |
|
not modelled |
23.5 |
15 |
PDB header:ligase
Chain: F: PDB Molecule:alpha-aminoadipate carrier protein lysw;
PDBTitle: argx from sulfolobus tokodaii complexed with2 lysw/glu/adp/mg/zn/sulfate
|
|
|
| 64 | c3lasA_ |
|
not modelled |
22.7 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:putative carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase from streptococcus mutans to2 1.4 angstrom resolution
|
|
|
| 65 | c5ztpB_ |
|
not modelled |
22.2 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: carbonic anhydrase from glaciozyma antarctica
|
|
|
| 66 | c2k5hA_ |
|
not modelled |
22.2 |
21 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved protein;
PDBTitle: solution nmr structure of protein encoded by mth693 from2 methanobacterium thermoautotrophicum: northeast structural3 genomics consortium target tt824a
|
|
|
| 67 | c2w3nA_ |
|
not modelled |
20.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase 2;
PDBTitle: structure and inhibition of the co2-sensing carbonic anhydrase can22 from the pathogenic fungus cryptococcus neoformans
|
|
|
| 68 | d2exda1 |
|
not modelled |
20.9 |
22 |
Fold:OB-fold
Superfamily:NfeD domain-like
Family:NfeD domain-like |
|
|
| 69 | d1ddza2 |
|
not modelled |
19.8 |
21 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
|
|
| 70 | c6gwuB_ |
|
not modelled |
19.4 |
8 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: carbonic anhydrase cance103p from candida albicans
|
|
|
| 71 | c6g4wr_ |
|
not modelled |
18.5 |
22 |
PDB header:ribosome
Chain: R: PDB Molecule:40s ribosomal protein s17;
PDBTitle: cryo-em structure of a late human pre-40s ribosomal subunit - state a
|
|
|
| 72 | c3cp0A_ |
|
not modelled |
18.4 |
17 |
PDB header:membrane protein
Chain: A: PDB Molecule:membrane protein implicated in regulation of membrane
PDBTitle: crystal structure of the soluble domain of membrane protein implicated2 in regulation of membrane protease activity from corynebacterium3 glutamicum
|
|
|
| 73 | c2a8cE_ |
|
not modelled |
18.2 |
17 |
PDB header:lyase
Chain: E: PDB Molecule:carbonic anhydrase 2;
PDBTitle: haemophilus influenzae beta-carbonic anhydrase
|
|
|
| 74 | d1ddza1 |
|
not modelled |
18.0 |
17 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
|
|
| 75 | c1ylkA_ |
|
not modelled |
18.0 |
17 |
PDB header:unknown function
Chain: A: PDB Molecule:hypothetical protein rv1284/mt1322;
PDBTitle: crystal structure of rv1284 from mycobacterium tuberculosis in complex2 with thiocyanate
|
|
|
| 76 | c5cxkG_ |
|
not modelled |
18.0 |
17 |
PDB header:lyase
Chain: G: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of beta carbonic anhydrase from vibrio cholerae
|
|
|
| 77 | d2nn6g3 |
|
not modelled |
17.9 |
21 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
|
|
| 78 | c3eyxB_ |
|
not modelled |
17.7 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of carbonic anhydrase nce103 from2 saccharomyces cerevisiae
|
|
|
| 79 | c5swcE_ |
|
not modelled |
17.7 |
8 |
PDB header:lyase
Chain: E: PDB Molecule:carbonic anhydrase;
PDBTitle: the structure of the beta-carbonic anhydrase ccaa
|
|
|
| 80 | c1ddzA_ |
|
not modelled |
17.3 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: x-ray structure of a beta-carbonic anhydrase from the red2 alga, porphyridium purpureum r-1
|
|
|
| 81 | c3ucoB_ |
|
not modelled |
17.0 |
8 |
PDB header:lyase/lyase inhibitor
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: coccomyxa beta-carbonic anhydrase in complex with iodide
|
|
|
| 82 | c2a5vB_ |
|
not modelled |
16.7 |
33 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase (carbonate dehydratase) (carbonic
PDBTitle: crystal structure of m. tuberculosis beta carbonic anhydrase, rv3588c,2 tetrameric form
|
|
|
| 83 | d2aqaa1 |
|
not modelled |
16.5 |
20 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
|
|
| 84 | c2j6aA_ |
|
not modelled |
16.4 |
6 |
PDB header:transferase
Chain: A: PDB Molecule:protein trm112;
PDBTitle: crystal structure of s. cerevisiae ynr046w, a zinc finger2 protein from the erf1 methyltransferase complex.
|
|
|
| 85 | d1ekja_ |
|
not modelled |
16.0 |
17 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
|
|
| 86 | c5cm2M_ |
|
not modelled |
16.0 |
0 |
PDB header:transferase
Chain: M: PDB Molecule:trna methyltransferase activator subunit;
PDBTitle: insights into molecular plasticity in protein complexes from trm9-2 trm112 trna modifying enzyme crystal structure
|
|
|
| 87 | c4rxyA_ |
|
not modelled |
15.3 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structure of the beta carbonic anhydrase psca3 isolated from2 pseudomonas aeruginosa
|
|
|
| 88 | d1i6pa_ |
|
not modelled |
15.2 |
25 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
|
|
| 89 | d1g5ca_ |
|
not modelled |
14.4 |
8 |
Fold:Resolvase-like
Superfamily:beta-carbonic anhydrase, cab
Family:beta-carbonic anhydrase, cab |
|
|
| 90 | c6g5iy_ |
|
not modelled |
14.3 |
13 |
PDB header:ribosome
Chain: Y: PDB Molecule:40s ribosomal protein s24;
PDBTitle: cryo-em structure of a late human pre-40s ribosomal subunit - state r
|
|
|
| 91 | d2apob1 |
|
not modelled |
12.6 |
19 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
|
|
| 92 | c3j20R_ |
|
not modelled |
12.6 |
20 |
PDB header:ribosome
Chain: R: PDB Molecule:30s ribosomal protein s17p;
PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
|
|
|
| 93 | d2ey4e1 |
|
not modelled |
12.5 |
19 |
Fold:Rubredoxin-like
Superfamily:Nop10-like SnoRNP
Family:Nucleolar RNA-binding protein Nop10-like |
|
|
| 94 | c4o1kA_ |
|
not modelled |
12.4 |
25 |
PDB header:lyase
Chain: A: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structures of two tetrameric beta-carbonic anhydrases from the2 filamentous ascomycete sordaria macrospora.
|
|
|
| 95 | d2j7ja2 |
|
not modelled |
12.0 |
33 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
|
|
| 96 | c2jrrA_ |
|
not modelled |
11.8 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution nmr structure of q5lls5 from silicibacter2 pomeroyi. northeast structural genomics consortium target3 sir90
|
|
|
| 97 | c4o1jB_ |
|
not modelled |
11.6 |
17 |
PDB header:lyase
Chain: B: PDB Molecule:carbonic anhydrase;
PDBTitle: crystal structures of two tetrameric beta-carbonic anhydrases from the2 filamentous ascomycete sordaria macrospora.
|
|
|
| 98 | d2c42a2 |
|
not modelled |
11.2 |
0 |
Fold:Thiamin diphosphate-binding fold (THDP-binding)
Superfamily:Thiamin diphosphate-binding fold (THDP-binding)
Family:PFOR PP module |
|
|
| 99 | d1ubdc1 |
|
not modelled |
10.3 |
33 |
Fold:beta-beta-alpha zinc fingers
Superfamily:beta-beta-alpha zinc fingers
Family:Classic zinc finger, C2H2 |
|
|