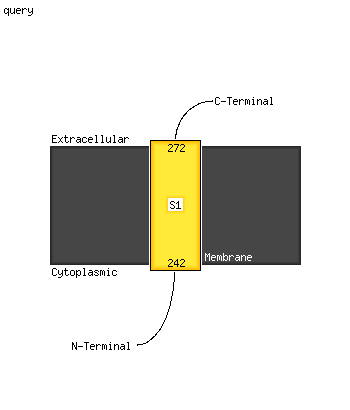
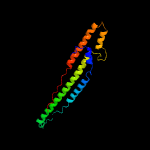
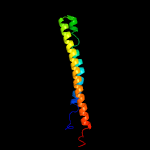
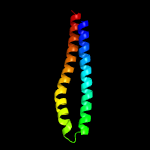
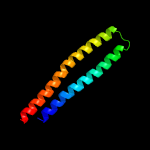
1 c5xfsB_
100.0
39
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
2 c2g38B_
100.0
30
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
3 d2g38b1
100.0
30
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
4 c4xy3A_
100.0
19
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion-associated protein espb;PDBTitle: structure of esx-1 secreted protein espb
5 c4wj2A_
97.9
20
PDB header: unknown functionChain: A: PDB Molecule: antigen mtb48;PDBTitle: mycobacterial protein
6 c2vs0B_
97.6
11
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
7 c4iogD_
97.4
14
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
8 c3gvmA_
97.3
14
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 c3zbhC_
97.1
15
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
10 d1wa8a1
96.4
18
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
11 c4lwsB_
93.5
18
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
12 c4lwsA_
93.3
12
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
13 d1wa8b1
92.9
17
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
14 c4i0xA_
91.4
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
15 c2kg7B_
86.0
19
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
16 c4i0xJ_
56.1
30
PDB header: structural genomics, unknown functionChain: J: PDB Molecule: esat-6-like protein mab_3113;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
17 d1ui5a2
45.0
22
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
18 d1zeea1
25.1
42
Fold: Indolic compounds 2,3-dioxygenase-likeSuperfamily: Indolic compounds 2,3-dioxygenase-likeFamily: Indoleamine 2,3-dioxygenase-like
19 c3zfsA_
24.8
18
PDB header: oxidoreductaseChain: A: PDB Molecule: f420-reducing hydrogenase, subunit alpha;PDBTitle: cryo-em structure of the f420-reducing nife-hydrogenase from a2 methanogenic archaeon with bound substrate
20 c4xb6D_
17.2
22
PDB header: transferaseChain: D: PDB Molecule: alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;PDBTitle: structure of the e. coli c-p lyase core complex
21 c5l85B_
not modelled
16.5
36
PDB header: signaling proteinChain: B: PDB Molecule: nuclear fragile x mental retardation-interacting protein 1;PDBTitle: solution structure of the complex between human znhit3 and nufip12 proteins
22 c3sjrB_
not modelled
16.4
20
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of conserved unkown function protein cv_1783 from2 chromobacterium violaceum atcc 12472
23 c2nvjA_
not modelled
15.8
40
PDB header: hydrolaseChain: A: PDB Molecule: 25mer peptide from vacuolar atp synthase subunitPDBTitle: nmr structures of transmembrane segment from subunit a from2 the yeast proton v-atpase
24 c2ahmG_
not modelled
15.7
29
PDB header: viral protein, replicationChain: G: PDB Molecule: replicase polyprotein 1ab, heavy chain;PDBTitle: crystal structure of sars-cov super complex of non-structural2 proteins: the hexadecamer
25 c3j00Z_
not modelled
14.6
4
PDB header: ribosome/ribosomal proteinChain: Z: PDB Molecule: cell division protein ftsq;PDBTitle: structure of the ribosome-secye complex in the membrane environment
26 d1dlpa1
not modelled
13.2
12
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins
27 c6aokA_
not modelled
12.7
38
PDB header: hydrolaseChain: A: PDB Molecule: ceg4;PDBTitle: crystal structure of legionella pneumophila effector ceg4 with n-2 terminal tev protease cleavage sequence
28 d1vlfn1
not modelled
12.3
100
Fold: Prealbumin-likeSuperfamily: Cna protein B-type domainFamily: Cna protein B-type domain
29 c1bkvA_
not modelled
11.6
44
PDB header: structural proteinChain: A: PDB Molecule: t3-785;PDBTitle: collagen
30 c6cgjA_
not modelled
11.4
38
PDB header: hydrolaseChain: A: PDB Molecule: effector protein lem4 (lpg1101);PDBTitle: structure of the had domain of effector protein lem4 (lpg1101) from2 legionella pneumophila
31 c1bkvC_
not modelled
11.2
44
PDB header: structural proteinChain: C: PDB Molecule: t3-785;PDBTitle: collagen
32 c1bkvB_
not modelled
11.2
44
PDB header: structural proteinChain: B: PDB Molecule: t3-785;PDBTitle: collagen
33 c3ub0D_
not modelled
10.8
17
PDB header: replicationChain: D: PDB Molecule: non-structural protein 6, nsp6,;PDBTitle: crystal structure of the nonstructural protein 7 and 8 complex of2 feline coronavirus
34 c2wseE_
not modelled
9.2
30
PDB header: photosynthesisChain: E: PDB Molecule: photosystem i reaction center subunit iv a, chloroplastic;PDBTitle: improved model of plant photosystem i
35 d1dmua_
not modelled
8.8
44
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Restriction endonuclease BglI
36 c1bzgA_
not modelled
8.8
14
PDB header: hormoneChain: A: PDB Molecule: parathyroid hormone-related protein;PDBTitle: the solution structure of human parathyroid hormone-related2 protein (1-34) in near-physiological solution, nmr, 303 structures
37 c2kg7A_
not modelled
8.8
59
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein esxg (pe family protein);PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
38 c4u39Q_
not modelled
8.6
36
PDB header: cell cycleChain: Q: PDB Molecule: cell division factor;PDBTitle: crystal structure of ftsz:mciz complex from bacillus subtilis
39 c4f3fC_
not modelled
8.5
38
PDB header: immune systemChain: C: PDB Molecule: mesothelin;PDBTitle: crystal structure of msln7-64 morab-009 fab complex
40 c5hl8B_
not modelled
8.2
40
PDB header: protein transportChain: B: PDB Molecule: type ii secretion system protein l;PDBTitle: 1.93 angstrom resolution crystal structure of a pullulanase-specific2 type ii secretion system integral cytoplasmic membrane protein gspl3 (c-terminal fragment; residues 309-397) from klebsiella pneumoniae4 subsp. pneumoniae ntuh-k2044
41 d1nkta4
not modelled
8.1
58
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: Tandem AAA-ATPase domain
42 c6nbiP_
not modelled
7.9
60
PDB header: signaling proteinChain: P: PDB Molecule: long-acting parathyroid hormone analog;PDBTitle: cryo-em structure of parathyroid hormone receptor type 1 in complex2 with a long-acting parathyroid hormone analog and g protein
43 c2m5lA_
not modelled
7.6
50
PDB header: viral proteinChain: A: PDB Molecule: ns5a protein;PDBTitle: ns5a308
44 c1vytF_
not modelled
7.5
50
PDB header: transport proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channelPDBTitle: beta3 subunit complexed with aid
45 c5dn4A_
not modelled
7.0
43
PDB header: hydrolaseChain: A: PDB Molecule: peptidoglycan hydrolase flgj;PDBTitle: structure of the glycoside hydrolase domain from salmonella2 typhimurium flgj
46 c4gyxC_
not modelled
6.9
38
PDB header: structural protein, blood clottingChain: C: PDB Molecule: type iii collagen fragment in a host peptide stabilized byPDBTitle: the von willebrand factor a3 domain binding region of type iii2 collagen stabilized by the cysteine knot
47 c2jtwA_
not modelled
6.9
50
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane helix 7 of yeast vatpase;PDBTitle: solution structure of tm7 bound to dpc micelles
48 c3ogrA_
not modelled
6.8
38
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: complex structure of beta-galactosidase from trichoderma reesei with2 galactose
49 d1fcda3
not modelled
6.8
40
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
50 c6fxoA_
not modelled
6.8
67
PDB header: hydrolaseChain: A: PDB Molecule: bifunctional autolysin;PDBTitle: crystal structure of major bifunctional autolysin
51 d2apla1
not modelled
6.4
32
Fold: PG0816-likeSuperfamily: PG0816-likeFamily: PG0816-like
52 c6gp2A_
not modelled
6.4
12
PDB header: oxidoreductaseChain: A: PDB Molecule: ribonucleoside-diphosphate reductase beta chain;PDBTitle: ribonucleotide reductase class ie r2 from mesoplasma florum, dopa-2 active form
53 c2lkqA_
not modelled
6.4
56
PDB header: immune systemChain: A: PDB Molecule: immunoglobulin lambda-like polypeptide 1;PDBTitle: nmr structure of the lambda 5 22-45 peptide
54 c3e6qL_
not modelled
6.3
20
PDB header: isomeraseChain: L: PDB Molecule: putative 5-carboxymethyl-2-hydroxymuconate isomerase;PDBTitle: putative 5-carboxymethyl-2-hydroxymuconate isomerase from pseudomonas2 aeruginosa.
55 c4dmtB_
not modelled
6.3
38
PDB header: structural proteinChain: B: PDB Molecule: collagen iii derived peptide;PDBTitle: crystal structure of a vwf binding collagen iii derived triple helical2 peptide
56 c4dmtC_
not modelled
6.3
38
PDB header: structural proteinChain: C: PDB Molecule: collagen iii derived peptide;PDBTitle: crystal structure of a vwf binding collagen iii derived triple helical2 peptide
57 c4dmtA_
not modelled
6.3
38
PDB header: structural proteinChain: A: PDB Molecule: collagen iii derived peptide;PDBTitle: crystal structure of a vwf binding collagen iii derived triple helical2 peptide
58 c2iunD_
not modelled
6.2
50
PDB header: viral proteinChain: D: PDB Molecule: avian adenovirus celo long fibre;PDBTitle: structure of the c-terminal head domain of the avian adenovirus celo2 long fibre (p21 crystal form)
59 c4cuaB_
not modelled
6.2
14
PDB header: hydrolaseChain: B: PDB Molecule: beta-galactosidase;PDBTitle: unravelling the multiple functions of the architecturally2 intricate streptococcus pneumoniae beta-galactosidase, bgaa
60 c2zycA_
not modelled
6.0
60
PDB header: hydrolaseChain: A: PDB Molecule: peptidoglycan hydrolase flgj;PDBTitle: crystal structure of peptidoglycan hydrolase from2 sphingomonas sp. a1
61 c1t0jC_
not modelled
6.0
43
PDB header: signaling proteinChain: C: PDB Molecule: voltage-dependent l-type calcium channel alpha-1c subunit;PDBTitle: crystal structure of a complex between voltage-gated calcium channel2 beta2a subunit and a peptide of the alpha1c subunit
62 c5uc0B_
not modelled
6.0
100
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein cog5400;PDBTitle: crystal structure of beta-barrel-like, uncharacterized protein of2 cog5400 from brucella abortus
63 c6qpiA_
not modelled
5.9
19
PDB header: membrane proteinChain: A: PDB Molecule: anoctamin-6;PDBTitle: cryo-em structure of calcium-free mtmem16f lipid scramblase in2 nanodisc
64 c4mveB_
not modelled
5.8
24
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of tcur_1030 protein from thermomonospora curvata
65 c1vytE_
not modelled
5.8
50
PDB header: transport proteinChain: E: PDB Molecule: voltage-dependent l-type calcium channelPDBTitle: beta3 subunit complexed with aid
66 c2d9zA_
not modelled
5.7
35
PDB header: signaling proteinChain: A: PDB Molecule: protein kinase c, nu type;PDBTitle: solution structure of the ph domain of protein kinase c, nu2 type from human
67 c4gyxA_
not modelled
5.7
45
PDB header: structural protein, blood clottingChain: A: PDB Molecule: type iii collagen fragment in a host peptide stabilized byPDBTitle: the von willebrand factor a3 domain binding region of type iii2 collagen stabilized by the cysteine knot
68 c4gyxB_
not modelled
5.7
45
PDB header: structural protein, blood clottingChain: B: PDB Molecule: type iii collagen fragment in a host peptide stabilized byPDBTitle: the von willebrand factor a3 domain binding region of type iii2 collagen stabilized by the cysteine knot
69 c6et5U_
not modelled
5.7
58
PDB header: photosynthesisChain: U: PDB Molecule: light-harvesting protein b-1015 gamma chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
70 c6et5g_
not modelled
5.7
58
PDB header: photosynthesisChain: G: PDB Molecule: light-harvesting protein b-1015 beta chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
71 c6et5d_
not modelled
5.7
58
PDB header: photosynthesisChain: D: PDB Molecule: PDBTitle: reaction centre light harvesting complex 1 from blc. virids
72 c6et5I_
not modelled
5.7
58
PDB header: photosynthesisChain: I: PDB Molecule: light-harvesting protein b-1015 gamma chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
73 c6et5R_
not modelled
5.7
58
PDB header: photosynthesisChain: R: PDB Molecule: light-harvesting protein b-1015 gamma chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
74 c6et5s_
not modelled
5.7
58
PDB header: photosynthesisChain: S: PDB Molecule: light-harvesting protein b-1015 alpha chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
75 c6et5a_
not modelled
5.7
58
PDB header: photosynthesisChain: A: PDB Molecule: PDBTitle: reaction centre light harvesting complex 1 from blc. virids
76 c6et5v_
not modelled
5.7
58
PDB header: photosynthesisChain: V: PDB Molecule: light-harvesting protein b-1015 alpha chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
77 c6et5m_
not modelled
5.7
58
PDB header: photosynthesisChain: M: PDB Molecule: reaction center protein m chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
78 c6et5p_
not modelled
5.7
58
PDB header: photosynthesisChain: P: PDB Molecule: light-harvesting protein b-1015 alpha chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
79 c6et52_
not modelled
5.7
58
PDB header: photosynthesisChain: 2: PDB Molecule: light-harvesting protein b-1015 gamma chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
80 c6et5O_
not modelled
5.7
58
PDB header: photosynthesisChain: O: PDB Molecule: light-harvesting protein b-1015 gamma chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
81 c6et5j_
not modelled
5.7
58
PDB header: photosynthesisChain: J: PDB Molecule: PDBTitle: reaction centre light harvesting complex 1 from blc. virids
82 c6et5y_
not modelled
5.7
58
PDB header: photosynthesisChain: Y: PDB Molecule: light-harvesting protein b-1015 alpha chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
83 c6et5X_
not modelled
5.7
58
PDB header: photosynthesisChain: X: PDB Molecule: light-harvesting protein b-1015 gamma chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
84 c6et55_
not modelled
5.7
58
PDB header: photosynthesisChain: 5: PDB Molecule: light-harvesting protein b-1015 gamma chain;PDBTitle: reaction centre light harvesting complex 1 from blc. virids
85 c4qdnA_
not modelled
5.7
80
PDB header: hydrolaseChain: A: PDB Molecule: flagellar protein flgj [peptidoglycan hydrolase];PDBTitle: crystal structure of the endo-beta-n-acetylglucosaminidase from2 thermotoga maritima
86 c2vwaE_
not modelled
5.6
25
PDB header: unknown functionChain: E: PDB Molecule: putative uncharacterized protein pf13_0012;PDBTitle: crystal structure of a sporozoite protein essential for2 liver stage development of malaria parasite
87 c6dzsD_
not modelled
5.5
31
PDB header: oxidoreductaseChain: D: PDB Molecule: homoserine dehydrogenase;PDBTitle: mycobacterial homoserine dehydrogenase thra in complex with nadp
88 c3qi7A_
not modelled
5.5
57
PDB header: transcriptionChain: A: PDB Molecule: putative transcriptional regulator;PDBTitle: crystal structure of a putative transcriptional regulator2 (yp_001089212.1) from clostridium difficile 630 at 1.86 a resolution
89 c4dexB_
not modelled
5.5
50
PDB header: transport proteinChain: B: PDB Molecule: voltage-dependent n-type calcium channel subunit alpha-1b;PDBTitle: crystal structure of the voltage dependent calcium channel beta-22 subunit in complex with the cav2.2 i-ii linker.
90 d1otga_
not modelled
5.5
23
Fold: Tautomerase/MIFSuperfamily: Tautomerase/MIFFamily: 5-carboxymethyl-2-hydroxymuconate isomerase (CHMI)
91 c1e1hD_
not modelled
5.4
60
PDB header: hydrolaseChain: D: PDB Molecule: botulinum neurotoxin type a light chain;PDBTitle: crystal structure of recombinant botulinum neurotoxin type a light2 chain, self-inhibiting zn endopeptidase.
92 c6igzE_
not modelled
5.4
31
PDB header: plant proteinChain: E: PDB Molecule: psae;PDBTitle: structure of psi-lhci
93 c4lzxB_
not modelled
5.3
31
PDB header: metal binding proteinChain: B: PDB Molecule: iq domain-containing protein g;PDBTitle: complex of iqcg and ca2+-free cam
94 c5vmoB_
not modelled
5.3
50
PDB header: viral protein/apoptosisChain: B: PDB Molecule: bcl-2 interacting mediator of cell death;PDBTitle: crystal structure of grouper iridovirus giv66:bim complex
95 c3fi7A_
not modelled
5.3
80
PDB header: hydrolaseChain: A: PDB Molecule: lmo1076 protein;PDBTitle: crystal structure of the autolysin auto (lmo1076) from listeria2 monocytogenes, catalytic domain
96 c4m1lB_
not modelled
5.2
36
PDB header: metal binding proteinChain: B: PDB Molecule: iq domain-containing protein g;PDBTitle: complex of iqcg and ca2+-bound cam
97 d1jyaa_
not modelled
5.2
19
Fold: Secretion chaperone-likeSuperfamily: Type III secretory system chaperone-likeFamily: Type III secretory system chaperone
98 c2o01E_
not modelled
5.1
38
PDB header: photosynthesisChain: E: PDB Molecule: photosystem i reaction center subunit iv a, chloroplast;PDBTitle: the structure of a plant photosystem i supercomplex at 3.4 angstrom2 resolution