| 1 |
|
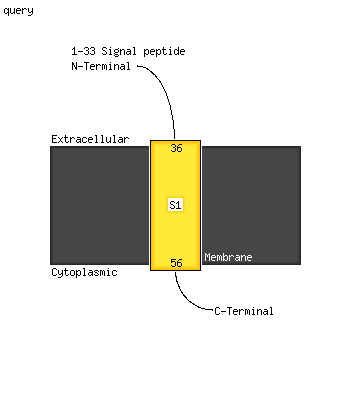
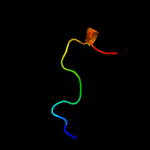
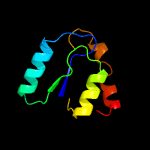
PDB 2k42 chain B
Region: 163 - 176
Aligned: 14
Modelled: 14
Confidence: 33.6%
Identity: 50%
PDB header:signaling protein
Chain: B: PDB Molecule:espfu;
PDBTitle: solution structure of the gtpase binding domain of wasp in2 complex with espfu, an ehec effector
Phyre2
| 2 |
|
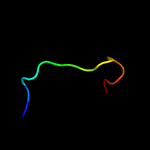
PDB 2lnh chain C
Region: 163 - 176
Aligned: 14
Modelled: 14
Confidence: 28.7%
Identity: 50%
PDB header:signaling protein/protein binding
Chain: C: PDB Molecule:secreted effector protein espf(u);
PDBTitle: enterohaemorrhagic e. coli (ehec) exploits a tryptophan switch to2 hijack host f-actin assembly
Phyre2
| 3 |
|
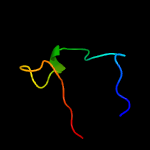
PDB 2iea chain A domain 3
Region: 98 - 119
Aligned: 22
Modelled: 22
Confidence: 12.9%
Identity: 18%
Fold: TK C-terminal domain-like
Superfamily: TK C-terminal domain-like
Family: Transketolase C-terminal domain-like
Phyre2
| 4 |
|
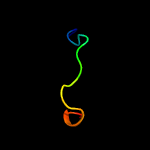
PDB 5cdj chain A
Region: 200 - 215
Aligned: 15
Modelled: 16
Confidence: 10.9%
Identity: 33%
PDB header:chaperone
Chain: A: PDB Molecule:rubisco large subunit-binding protein subunit alpha,
PDBTitle: apical domain of chloroplast chaperonin 60a
Phyre2
| 5 |
|
PDB 2lgx chain A
Region: 108 - 117
Aligned: 10
Modelled: 10
Confidence: 10.3%
Identity: 70%
PDB header:cell adhesion
Chain: A: PDB Molecule:fermitin family homolog 2;
PDBTitle: nmr structure for kindle-2 n-terminus
Phyre2
| 6 |
|
PDB 2lzs chain E
Region: 35 - 50
Aligned: 16
Modelled: 16
Confidence: 10.3%
Identity: 38%
PDB header:protein transport
Chain: E: PDB Molecule:sec-independent protein translocase protein tata;
PDBTitle: tata oligomer
Phyre2
| 7 |
|
PDB 1sjp chain A domain 2
Region: 200 - 215
Aligned: 15
Modelled: 16
Confidence: 10.2%
Identity: 47%
Fold: The "swivelling" beta/beta/alpha domain
Superfamily: GroEL apical domain-like
Family: GroEL-like chaperone, apical domain
Phyre2
| 8 |
|
PDB 1kid chain A
Region: 196 - 215
Aligned: 19
Modelled: 20
Confidence: 10.1%
Identity: 26%
Fold: The "swivelling" beta/beta/alpha domain
Superfamily: GroEL apical domain-like
Family: GroEL-like chaperone, apical domain
Phyre2
| 9 |
|
PDB 1iok chain A domain 2
Region: 200 - 215
Aligned: 15
Modelled: 16
Confidence: 9.5%
Identity: 40%
Fold: The "swivelling" beta/beta/alpha domain
Superfamily: GroEL apical domain-like
Family: GroEL-like chaperone, apical domain
Phyre2
| 10 |
|
PDB 1dk7 chain A
Region: 200 - 215
Aligned: 15
Modelled: 16
Confidence: 9.3%
Identity: 33%
Fold: The "swivelling" beta/beta/alpha domain
Superfamily: GroEL apical domain-like
Family: GroEL-like chaperone, apical domain
Phyre2
| 11 |
|
PDB 1srv chain A
Region: 200 - 226
Aligned: 26
Modelled: 27
Confidence: 9.3%
Identity: 23%
Fold: The "swivelling" beta/beta/alpha domain
Superfamily: GroEL apical domain-like
Family: GroEL-like chaperone, apical domain
Phyre2
| 12 |
|
PDB 1oel chain A domain 2
Region: 200 - 215
Aligned: 15
Modelled: 16
Confidence: 9.0%
Identity: 33%
Fold: The "swivelling" beta/beta/alpha domain
Superfamily: GroEL apical domain-like
Family: GroEL-like chaperone, apical domain
Phyre2
| 13 |
|
PDB 1we3 chain A domain 2
Region: 200 - 215
Aligned: 15
Modelled: 16
Confidence: 8.4%
Identity: 33%
Fold: The "swivelling" beta/beta/alpha domain
Superfamily: GroEL apical domain-like
Family: GroEL-like chaperone, apical domain
Phyre2
| 14 |
|
PDB 3m6c chain A
Region: 200 - 226
Aligned: 26
Modelled: 27
Confidence: 8.2%
Identity: 23%
PDB header:chaperone
Chain: A: PDB Molecule:60 kda chaperonin 1;
PDBTitle: crystal structure of mycobacterium tuberculosis groel1 apical domain
Phyre2
| 15 |
|
PDB 2i2x chain D
Region: 87 - 181
Aligned: 79
Modelled: 79
Confidence: 7.8%
Identity: 25%
PDB header:transferase
Chain: D: PDB Molecule:methyltransferase 1;
PDBTitle: crystal structure of methanol:cobalamin methyltransferase complex2 mtabc from methanosarcina barkeri
Phyre2
| 16 |
|
PDB 2kmc chain A
Region: 108 - 116
Aligned: 9
Modelled: 9
Confidence: 7.1%
Identity: 56%
PDB header:cell adhesion
Chain: A: PDB Molecule:fermitin family homolog 1;
PDBTitle: solution structure of the n-terminal domain of kindlin-1
Phyre2
| 17 |
|
PDB 1zj8 chain A domain 2
Region: 86 - 123
Aligned: 36
Modelled: 38
Confidence: 6.7%
Identity: 17%
Fold: Ferredoxin-like
Superfamily: Nitrite/Sulfite reductase N-terminal domain-like
Family: Duplicated SiR/NiR-like domains 1 and 3
Phyre2
| 18 |
|
PDB 2hjj chain A domain 1
Region: 104 - 116
Aligned: 13
Modelled: 13
Confidence: 6.2%
Identity: 46%
Fold: dsRBD-like
Superfamily: YcfA/nrd intein domain
Family: YkfF-like
Phyre2
| 19 |
|
PDB 2hjj chain A
Region: 104 - 116
Aligned: 13
Modelled: 13
Confidence: 6.2%
Identity: 46%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ykff;
PDBTitle: solution nmr structure of protein ykff from escherichia coli.2 northeast structural genomics target er397.
Phyre2
| 20 |
|
PDB 1ccw chain A
Region: 87 - 177
Aligned: 75
Modelled: 75
Confidence: 5.9%
Identity: 19%
Fold: Flavodoxin-like
Superfamily: Cobalamin (vitamin B12)-binding domain
Family: Cobalamin (vitamin B12)-binding domain
Phyre2
| 21 |
|
| 22 |
|