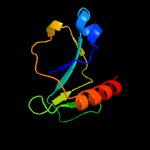
1 c5hk3B_
97.2
15
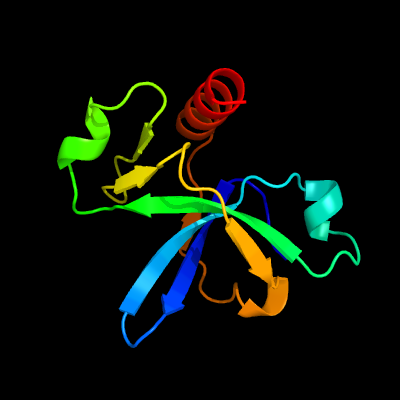
PDB header: hydrolase/dnaChain: B: PDB Molecule: endoribonuclease mazf6;PDBTitle: crystal structure of m. tuberculosis mazf-mt3 t52d-f62d mutant in2 complex with dna
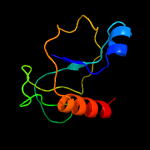
2 c5xe3B_
87.7
18
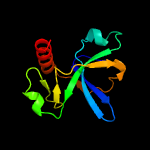
PDB header: hydrolase/antitoxinChain: B: PDB Molecule: endoribonuclease mazf4;PDBTitle: endoribonuclease in complex with its cognate antitoxin from2 mycobacterial species
3 c5wygC_
87.3
17
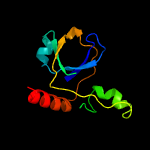
PDB header: hydrolaseChain: C: PDB Molecule: probable endoribonuclease mazf7;PDBTitle: the crystal structure of the apo form of mtb mazf
4 d1ub4a_
83.7
18
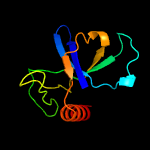
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
5 c5ccaA_
80.3
16
PDB header: hydrolaseChain: A: PDB Molecule: endoribonuclease mazf3;PDBTitle: crystal structure of mtb toxin
6 d1ne8a_
60.6
16
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
7 c4mzpC_
60.3
18
PDB header: hydrolaseChain: C: PDB Molecule: mazf mrna interferase;PDBTitle: mazf from s. aureus crystal form iii, c2221, 2.7 a
8 c5hjzA_
54.0
17
PDB header: hydrolase/rnaChain: A: PDB Molecule: endoribonuclease mazf9;PDBTitle: structure of m. tuberculosis mazf-mt1 (rv2801c) in complex with rna
9 d1m1fa_
26.4
14
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
10 c5a9qZ_
25.2
43
PDB header: transport proteinChain: Z: PDB Molecule: nuclear pore complex protein nup85;PDBTitle: human nuclear pore complex
11 c6g90J_
18.2
35
PDB header: splicingChain: J: PDB Molecule: u1 small nuclear ribonucleoprotein component snu71;PDBTitle: prespliceosome structure provides insight into spliceosome assembly2 and regulation (map a2)
12 c3j6vL_
17.3
67
PDB header: ribosomeChain: L: PDB Molecule: 28s ribosomal protein s12, mitochondrial;PDBTitle: cryo-em structure of the small subunit of the mammalian mitochondrial2 ribosome
13 c6fblA_
15.7
29
PDB header: rna binding proteinChain: A: PDB Molecule: mina-1;PDBTitle: nmr solution structure of mina-1(254-334)
14 c1zn1L_
15.7
33
PDB header: biosynthetic/structural protein/rnaChain: L: PDB Molecule: 30s ribosomal protein s12;PDBTitle: coordinates of rrf fitted into cryo-em map of the 70s post-termination2 complex
15 d2uubl1
15.4
42
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
16 d1i94l_
15.3
42
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
17 d2qall1
14.2
33
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
18 c1s1hL_
14.0
33
PDB header: ribosomeChain: L: PDB Molecule: 40s ribosomal protein s23;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1h, contains 40s subunit. the 60s4 ribosomal subunit is in file 1s1i.
19 c5xyiX_
13.6
33
PDB header: ribosomeChain: X: PDB Molecule: 40s ribosomal protein s23, putative;PDBTitle: small subunit of trichomonas vaginalis ribosome
20 c3u5gX_
13.5
33
PDB header: ribosomeChain: X: PDB Molecule: 40s ribosomal protein s23-a;PDBTitle: the structure of the eukaryotic ribosome at 3.0 a resolution. this2 entry contains proteins of the 40s subunit, ribosome b
21 c3zeyS_
not modelled
13.1
25
PDB header: ribosomeChain: S: PDB Molecule: 40s ribosomal protein s23, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
22 c3j20N_
not modelled
13.0
50
PDB header: ribosomeChain: N: PDB Molecule: 30s ribosomal protein s12p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (30s3 ribosomal subunit)
23 c2zkql_
not modelled
13.0
42
PDB header: ribosomal protein/rnaChain: L: PDB Molecule: PDBTitle: structure of a mammalian ribosomal 40s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
24 c3h7yA_
not modelled
12.1
15
PDB header: biosynthetic proteinChain: A: PDB Molecule: bacilysin biosynthesis protein bacb;PDBTitle: crystal structure of bacb, an enzyme involved in bacilysin synthesis,2 in tetragonal form
25 c2xzmL_
not modelled
11.5
25
PDB header: ribosomeChain: L: PDB Molecule: 40s ribosomal protein s12;PDBTitle: crystal structure of the eukaryotic 40s ribosomal2 subunit in complex with initiation factor 1. this file3 contains the 40s subunit and initiation factor for4 molecule 1
26 c1meqA_
not modelled
8.1
38
PDB header: viral proteinChain: A: PDB Molecule: exterior membrane glycoprotein (gp120);PDBTitle: hiv gp120 c5
27 d1t98a1
not modelled
7.9
83
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: MukF N-terminal domain-like
28 c6roiC_
not modelled
7.5
26
PDB header: lipid transportChain: C: PDB Molecule: cell division control protein 50;PDBTitle: cryo-em structure of the partially activated drs2p-cdc50p
29 d2es7a1
not modelled
7.0
25
Fold: Thioredoxin foldSuperfamily: Thioredoxin-likeFamily: HyaE-like
30 c5vbaA_
not modelled
7.0
23
PDB header: chaperone, hydrolaseChain: A: PDB Molecule: lysozyme, esx-1 secretion-associated protein espg1 chimera;PDBTitle: structure of espg1 chaperone from the type vii (esx-1) secretion2 system determined with the assistance of n-terminal t4 lysozyme3 fusion
31 c2kpoA_
not modelled
6.0
56
PDB header: de novo proteinChain: A: PDB Molecule: rossmann 2x2 fold protein;PDBTitle: solution nmr structure of de novo designed rossmann 2x2 fold protein,2 northeast structural genomics consortium target or16
32 c6fcxA_
not modelled
5.9
26
PDB header: oxidoreductaseChain: A: PDB Molecule: methylenetetrahydrofolate reductase;PDBTitle: structure of human 5,10-methylenetetrahydrofolate reductase (mthfr)
33 c2lpeA_
not modelled
5.6
23
PDB header: signaling proteinChain: A: PDB Molecule: kinase suppressor of ras 1;PDBTitle: solution nmr structure of the ksr1 ca1-ca1a domain