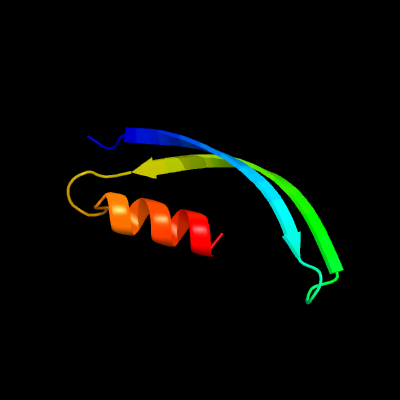
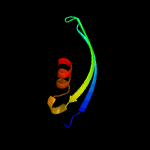
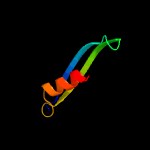
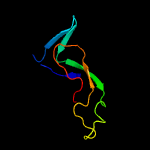
1 d2g3wa1
45.2
27
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: YaeQ-like
2 c3c0uA_
39.9
24
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein yaeq;PDBTitle: crystal structure of e.coli yaeq protein
3 d2ot9a1
39.1
20
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: YaeQ-like
4 c4ldxB_
25.8
29
PDB header: transcription/dnaChain: B: PDB Molecule: auxin response factor 1;PDBTitle: crystal structure of the dna binding domain of arabidopsis thaliana2 auxin response factor 1 (arf1) in complex with protomor-like sequence3 er7
5 c3zeyI_
12.0
33
PDB header: ribosomeChain: I: PDB Molecule: 40s ribosomal protein s15, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
6 d1t95a3
9.8
20
Fold: Ferredoxin-likeSuperfamily: EF-G C-terminal domain-likeFamily: Hypothetical protein AF0491, C-terminal domain
7 c3lgeF_
9.4
56
PDB header: lyase/protein bindingChain: F: PDB Molecule: sorting nexin-9;PDBTitle: crystal structure of rabbit muscle aldolase-snx9 lc4 complex
8 c3lgeG_
9.4
56
PDB header: lyase/protein bindingChain: G: PDB Molecule: sorting nexin-9;PDBTitle: crystal structure of rabbit muscle aldolase-snx9 lc4 complex
9 c2m9uA_
9.4
29
PDB header: viral proteinChain: A: PDB Molecule: integrase p46;PDBTitle: solution nmr structure of the c-terminal domain (ctd) of moloney2 murine leukemia virus integrase, northeast structural genomics target3 or41a
10 d3bz6a1
9.2
58
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: "Winged helix" DNA-binding domainFamily: PSPTO2686-like
11 d1wjna_
8.7
30
Fold: beta-Grasp (ubiquitin-like)Superfamily: Ubiquitin-likeFamily: Ubiquitin-related
12 c5ze4A_
7.7
32
PDB header: lyaseChain: A: PDB Molecule: dihydroxy-acid dehydratase, chloroplastic;PDBTitle: the structure of holo- structure of dhad complex with [2fe-2s] cluster
13 c6gcsj_
7.6
67
PDB header: oxidoreductaseChain: J: PDB Molecule: nujm subunit;PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
14 c3gxvD_
6.3
70
PDB header: hydrolase/replicationChain: D: PDB Molecule: replicative dna helicase;PDBTitle: three-dimensional structure of n-terminal domain of dnab2 helicase from helicobacter pylori and its interactions with3 primase
15 c2qldA_
6.3
24
PDB header: chaperoneChain: A: PDB Molecule: dnaj homolog subfamily b member 1;PDBTitle: human hsp40 hdj1
16 c5ym0A_
6.1
30
PDB header: lyaseChain: A: PDB Molecule: dihydroxy-acid dehydratase, chloroplastic;PDBTitle: the crystal structure of dhad
17 c3gxvC_
6.0
70
PDB header: hydrolase/replicationChain: C: PDB Molecule: replicative dna helicase;PDBTitle: three-dimensional structure of n-terminal domain of dnab2 helicase from helicobacter pylori and its interactions with3 primase
18 d1c5ea_
5.8
55
Fold: beta-clipSuperfamily: Head decoration protein D (gpD, major capsid protein D)Family: Head decoration protein D (gpD, major capsid protein D)
19 d2a0ua1
5.6
18
Fold: NagB/RpiA/CoA transferase-likeSuperfamily: NagB/RpiA/CoA transferase-likeFamily: IF2B-like
20 c4wfdA_
5.5
5
PDB header: hydrolaseChain: A: PDB Molecule: exosome complex exonuclease rrp6;PDBTitle: structure of the rrp6-rrp47-mtr4 interaction
21 d1td4a_
not modelled
5.5
55
Fold: beta-clipSuperfamily: Head decoration protein D (gpD, major capsid protein D)Family: Head decoration protein D (gpD, major capsid protein D)
22 d1yq2a4
not modelled
5.5
19
Fold: SupersandwichSuperfamily: Galactose mutarotase-likeFamily: beta-Galactosidase, domain 5
23 c2ro5B_
not modelled
5.4
35
PDB header: transcriptionChain: B: PDB Molecule: stage v sporulation protein t;PDBTitle: rdc-refined solution structure of the n-terminal dna recognition2 domain of the bacillus subtilis transition-state regulator spovt
24 c1h7dA_
not modelled
5.2
50
PDB header: acyltransferaseChain: A: PDB Molecule: aminolevulinic acid synthase 2, erythroid;PDBTitle: solution structure of the 49 aa presequence of 5-alas