| 1 | d1a2za_
|
|
 |
100.0 |
35 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
|
|
|
| 2 | c4gxhC_
|
|
 |
100.0 |
39 |
PDB header:hydrolase
Chain: C: PDB Molecule:pyrrolidone-carboxylate peptidase;
PDBTitle: crystal structure of a pyrrolidone-carboxylate peptidase 1 (target id2 nysgrc-012831) from xenorhabdus bovienii ss-2004
|
|
|
|
| 3 | d1iofa_
|
|
 |
100.0 |
37 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
|
|
|
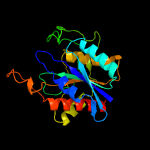
| 4 | d1auga_
|
|
 |
100.0 |
36 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
|
|
|
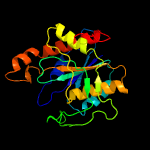
| 5 | c3giuA_
|
|
 |
100.0 |
30 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrrolidone-carboxylate peptidase;
PDBTitle: 1.25 angstrom crystal structure of pyrrolidone-carboxylate peptidase2 (pcp) from staphylococcus aureus
|
|
|
|
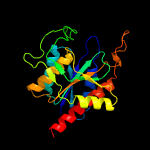
| 6 | d1iu8a_
|
|
 |
100.0 |
37 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase)
Family:Pyrrolidone carboxyl peptidase (pyroglutamate aminopeptidase) |
|
|
|
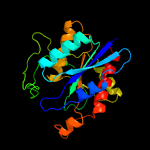
| 7 | c5z47A_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrrolidone-carboxylate peptidase;
PDBTitle: crystal structure of pyrrolidone carboxylate peptidase i with2 disordered loop a from deinococcus radiodurans r1
|
|
|
|
| 8 | c3lacA_
|
|
 |
100.0 |
38 |
PDB header:hydrolase
Chain: A: PDB Molecule:pyrrolidone-carboxylate peptidase;
PDBTitle: crystal structure of bacillus anthracis pyrrolidone-carboxylate2 peptidase, pcp
|
|
|
|
| 9 | c2ebjB_
|
|
 |
100.0 |
32 |
PDB header:hydrolase
Chain: B: PDB Molecule:pyrrolidone carboxyl peptidase;
PDBTitle: crystal structure of pyrrolidone carboxyl peptidase from thermus2 thermophilus
|
|
|
|
| 10 | c3wmwB_
|
|
 |
86.8 |
20 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nad dependent epimerase/dehydratase;
PDBTitle: gale-like l-threonine dehydrogenase from cupriavidus necator (apo2 form)
|
|
|
|
| 11 | c4b8wB_
|
|
 |
84.6 |
17 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-l-fucose synthase;
PDBTitle: crystal structure of human gdp-l-fucose synthase with bound nadp and2 gdp, tetragonal crystal form
|
|
|
|
| 12 | c2i5bC_
|
|
 |
79.6 |
17 |
PDB header:transferase
Chain: C: PDB Molecule:phosphomethylpyrimidine kinase;
PDBTitle: the crystal structure of an adp complex of bacillus subtilis pyridoxal2 kinase provides evidence for the parralel emergence of enzyme3 activity during evolution
|
|
|
|
| 13 | c3sc6F_
|
|
 |
79.3 |
22 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 2.65 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase (rfbd) from bacillus anthracis str. ames in complex with3 nadp
|
|
|
|
| 14 | c4wpgA_
|
|
 |
78.5 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: group a streptococcus gaca is an essential dtdp-4-dehydrorhamnose2 reductase (rmld)
|
|
|
|
| 15 | c2ydyA_
|
|
 |
76.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:methionine adenosyltransferase 2 subunit beta;
PDBTitle: crystal structure of human s-adenosylmethionine synthetase 2, beta2 subunit in orthorhombic crystal form
|
|
|
|
| 16 | d1jlja_
|
|
 |
71.5 |
20 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
|
| 17 | c6aqyD_
|
|
 |
68.9 |
23 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:gdp-l-fucose synthetase;
PDBTitle: crystal structure of a gdp-l-fucose synthetase from naegleria fowleri
|
|
|
|
| 18 | d1db3a_
|
|
 |
67.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
|
| 19 | c4josA_
|
|
 |
63.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteine nucleosidase;
PDBTitle: crystal structure of a putative 5'-methylthioadenosine/s-2 adenosylhomocysteine nucleosidase from francisella philomiragia atcc3 25017 (target nysgrc-029335)
|
|
|
|
| 20 | d2f7wa1
|
|
 |
62.6 |
9 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
|
| 21 | c1z45A_ |
|
not modelled |
61.5 |
26 |
PDB header:isomerase
Chain: A: PDB Molecule:gal10 bifunctional protein;
PDBTitle: crystal structure of the gal10 fusion protein galactose2 mutarotase/udp-galactose 4-epimerase from saccharomyces cerevisiae3 complexed with nad, udp-glucose, and galactose
|
|
|
| 22 | c2qq1A_ |
|
not modelled |
60.7 |
14 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdenum cofactor biosynthesis mog;
PDBTitle: crystal structure of molybdenum cofactor biosynthesis2 (aq_061) other form from aquifex aeolicus vf5
|
|
|
| 23 | d1n2sa_ |
|
not modelled |
59.7 |
10 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 24 | c4jgbB_ |
|
not modelled |
56.8 |
38 |
PDB header:protein binding
Chain: B: PDB Molecule:putative exported protein;
PDBTitle: crystal structure of putative exported protein from burkholderia2 pseudomallei
|
|
|
| 25 | d1bxka_ |
|
not modelled |
56.7 |
63 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 26 | c5b6kA_ |
|
not modelled |
55.3 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:uncharacterized protein cgkr1;
PDBTitle: crystal strucutre of ketoreductase 1 from candida glabrata
|
|
|
| 27 | c4dqvA_ |
|
not modelled |
55.1 |
22 |
PDB header:ligase
Chain: A: PDB Molecule:probable peptide synthetase nrp (peptide synthase);
PDBTitle: crystal structure of reductase (r) domain of non-ribosomal peptide2 synthetase from mycobacterium tuberculosis
|
|
|
| 28 | d1jysa_ |
|
not modelled |
54.8 |
16 |
Fold:Phosphorylase/hydrolase-like
Superfamily:Purine and uridine phosphorylases
Family:Purine and uridine phosphorylases |
|
|
| 29 | d1vl0a_ |
|
not modelled |
54.4 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 30 | c2is8A_ |
|
not modelled |
54.2 |
13 |
PDB header:structural protein
Chain: A: PDB Molecule:molybdopterin biosynthesis enzyme, moab;
PDBTitle: crystal structure of the molybdopterin biosynthesis enzyme moab2 (ttha0341) from thermus theromophilus hb8
|
|
|
| 31 | c5l9aB_ |
|
not modelled |
54.0 |
75 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:l-threonine 3-dehydrogenase;
PDBTitle: l-threonine dehydrogenase from trypanosoma brucei.
|
|
|
| 32 | c2q1wC_ |
|
not modelled |
54.0 |
63 |
PDB header:sugar binding protein
Chain: C: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmh in2 complex with nad+
|
|
|
| 33 | c3rfxB_ |
|
not modelled |
52.6 |
63 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:uronate dehydrogenase;
PDBTitle: crystal structure of uronate dehydrogenase from agrobacterium2 tumefaciens, y136a mutant complexed with nad
|
|
|
| 34 | d1oc2a_ |
|
not modelled |
51.3 |
38 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 35 | c3dhnA_ |
|
not modelled |
51.2 |
25 |
PDB header:isomerase, lyase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of the putative epimerase q89z24_bactn from2 bacteroides thetaiotaomicron. northeast structural genomics3 consortium target btr310.
|
|
|
| 36 | c4jwtA_ |
|
not modelled |
51.2 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:methylthioadenosine nucleosidase;
PDBTitle: crystal structure of a putative 5'-methylthioadenosine/s-2 adenosylhomocysteine nucleosidase from sulfurimonas denitrificans dsm3 1251 (target nysgrc-029304 )
|
|
|
| 37 | c4l0mA_ |
|
not modelled |
50.1 |
24 |
PDB header:hydrolase
Chain: A: PDB Molecule:putative 5'-methylthioadenosine/s-adenosylhomocysteine
PDBTitle: crystal structure of a putative 5'-methylthioadenosine/s-2 adenosylhomocysteine nucleosidase from borrelia burgdorferi b31 bound3 to adenine (target nysgrc-029268 )
|
|
|
| 38 | c4kn5A_ |
|
not modelled |
49.9 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:methylthioadenosine nucleosidase;
PDBTitle: crystal structure of a putative methylthioadenosine nucleosidase from2 weissella paramesenteroides atcc 33313 (target nysgrc-029342 )
|
|
|
| 39 | c6po4A_ |
|
not modelled |
48.1 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase;
PDBTitle: 2.1 angstrom resolution crystal structure of 5'-methylthioadenosine/s-2 adenosylhomocysteine nucleosidase (mtnn) from haemophilus influenzae3 pittii.
|
|
|
| 40 | d2c5aa1 |
|
not modelled |
47.9 |
38 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 41 | c5b7pB_ |
|
not modelled |
46.7 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:mta/sah nucleosidase;
PDBTitle: structures and functional analysis of periplasmic 5-2 methylthioadenosine/s-adenosylhomocysteine nucleosidase from3 aeromonas hydrophila
|
|
|
| 42 | c6g1nB_ |
|
not modelled |
46.3 |
14 |
PDB header:antitoxin
Chain: B: PDB Molecule:antitoxin hicb;
PDBTitle: crystal structure of the burkholderia pseudomallei antitoxin hicb
|
|
|
| 43 | c5wqnD_ |
|
not modelled |
45.1 |
50 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:probable dehydrogenase;
PDBTitle: crystal structure of a carbonyl reductase from pseudomonas aeruginosa2 pao1 (condition ii)
|
|
|
| 44 | c3c1oA_ |
|
not modelled |
44.0 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:eugenol synthase;
PDBTitle: the multiple phenylpropene synthases in both clarkia2 breweri and petunia hybrida represent two distinct lineages
|
|
|
| 45 | d1f0ka_ |
|
not modelled |
43.7 |
22 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Peptidoglycan biosynthesis glycosyltransferase MurG |
|
|
| 46 | c3ay3C_ |
|
not modelled |
43.0 |
50 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of glucuronic acid dehydrogeanse from2 chromohalobacter salexigens
|
|
|
| 47 | c3dp9A_ |
|
not modelled |
43.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:mta/sah nucleosidase;
PDBTitle: crystal structure of vibrio cholerae 5'-methylthioadenosine/s-adenosyl2 homocysteine nucleosidase (mtan) complexed with butylthio-dadme-3 immucillin a
|
|
|
| 48 | d1kewa_ |
|
not modelled |
41.9 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 49 | c4qezC_ |
|
not modelled |
41.8 |
20 |
PDB header:hydrolase
Chain: C: PDB Molecule:5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase;
PDBTitle: crystal structure of 5'-methylthioadenosine/s-adenosylhomocysteine2 nucleosidase from bacillus anthracis
|
|
|
| 50 | c1zosE_ |
|
not modelled |
41.5 |
14 |
PDB header:hydrolase
Chain: E: PDB Molecule:5'-methylthioadenosine / s-adenosylhomocysteine
PDBTitle: structure of 5'-methylthionadenosine/s-adenosylhomocysteine2 nucleosidase from s. pneumoniae with a transition-state inhibitor mt-3 imma
|
|
|
| 51 | c4bmvH_ |
|
not modelled |
40.8 |
50 |
PDB header:oxidoreductase
Chain: H: PDB Molecule:short-chain dehydrogenase;
PDBTitle: short-chain dehydrogenase from sphingobium yanoikuyae in2 complex with nadph
|
|
|
| 52 | c5msuC_ |
|
not modelled |
40.5 |
17 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:carboxylic acid reductase;
PDBTitle: structure of the r domain of carboxylic acid reductase (car) from2 mycobacterium marinum in complex with nadp, p21 form
|
|
|
| 53 | d2vbaa1 |
|
not modelled |
40.4 |
40 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
|
|
| 54 | c6if8D_ |
|
not modelled |
40.2 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase;
PDBTitle: aeromonas hydrophila mtan-2 complexed with adenine
|
|
|
| 55 | c3l6eA_ |
|
not modelled |
39.9 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:oxidoreductase, short-chain dehydrogenase/reductase family;
PDBTitle: crystal structure of putative short chain dehydrogenase/reductase2 family oxidoreductase from aeromonas hydrophila subsp. hydrophila3 atcc 7966
|
|
|
| 56 | c3gpiA_ |
|
not modelled |
39.9 |
50 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: structure of putative nad-dependent epimerase/dehydratase from2 methylobacillus flagellatus
|
|
|
| 57 | c1gsoA_ |
|
not modelled |
39.0 |
60 |
PDB header:ligase
Chain: A: PDB Molecule:protein (glycinamide ribonucleotide synthetase);
PDBTitle: glycinamide ribonucleotide synthetase (gar-syn) from e.2 coli.
|
|
|
| 58 | c5dk6A_ |
|
not modelled |
38.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase;
PDBTitle: crystal structure of a 5'-methylthioadenosine/s-adenosylhomocysteine2 (mta/sah) nucleosidase (mtan) from colwellia psychrerythraea 34h3 (cps_4743, target psi-029300) in complex with adenine at 2.27 a4 resolution
|
|
|
| 59 | c4qjiB_ |
|
not modelled |
37.2 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:phosphopantothenate--cysteine ligase;
PDBTitle: crystal structure of the c-terminal ctp-binding domain of a2 phosphopantothenoylcysteine decarboxylase/phosphopantothenate-3 cysteine ligase with bound ctp from mycobacterium smegmatis
|
|
|
| 60 | d1hdoa_ |
|
not modelled |
37.1 |
25 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 61 | d1di6a_ |
|
not modelled |
36.9 |
17 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 62 | c2b7lD_ |
|
not modelled |
36.1 |
25 |
PDB header:transferase
Chain: D: PDB Molecule:glycerol-3-phosphate cytidylyltransferase;
PDBTitle: crystal structure of ctp:glycerol-3-phosphate2 cytidylyltransferase from staphylococcus aureus
|
|
|
| 63 | d1e6ua_ |
|
not modelled |
36.0 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 64 | d2g2ca1 |
|
not modelled |
35.6 |
22 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 65 | c3eeiA_ |
|
not modelled |
35.4 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:5-methylthioadenosine nucleosidase/s-adenosylhomocysteine
PDBTitle: crystal structure of 5'-methylthioadenosine/s-adenosylhomocysteine2 nucleosidase from neisseria meningitidis in complex with methylthio-3 immucillin-a
|
|
|
| 66 | c5idwA_ |
|
not modelled |
34.9 |
50 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain dehydrogenase/reductase sdr;
PDBTitle: crystal structure of an oxidoreductase from burkholderia vietnamiensis2 in complex with nadp
|
|
|
| 67 | c3ia7A_ |
|
not modelled |
33.9 |
23 |
PDB header:transferase
Chain: A: PDB Molecule:calg4;
PDBTitle: crystal structure of calg4, the calicheamicin glycosyltransferase
|
|
|
| 68 | c4xcwF_ |
|
not modelled |
33.8 |
16 |
PDB header:transferase
Chain: F: PDB Molecule:molybdopterin adenylyltransferase;
PDBTitle: crystal structure of molybdenum cofactor biosynthesis protein moga2 from helicobacter pylori str. j99
|
|
|
| 69 | c4g41A_ |
|
not modelled |
33.7 |
9 |
PDB header:hydrolase
Chain: A: PDB Molecule:mta/sah nucleosidase;
PDBTitle: crystal structure of s-adenosylhomocysteine nucleosidase from2 streptococcus pyogenes in complex with 5-methylthiotubericidin
|
|
|
| 70 | c5u9cC_ |
|
not modelled |
33.5 |
19 |
PDB header:hydrolase,oxidoreductase
Chain: C: PDB Molecule:dtdp-4-dehydrorhamnose reductase;
PDBTitle: 1.9 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 reductase from yersinia enterocolitica
|
|
|
| 71 | c3eywA_ |
|
not modelled |
33.4 |
42 |
PDB header:transport protein
Chain: A: PDB Molecule:c-terminal domain of glutathione-regulated potassium-efflux
PDBTitle: crystal structure of the c-terminal domain of e. coli kefc in complex2 with keff
|
|
|
| 72 | c5yrzC_ |
|
not modelled |
33.2 |
14 |
PDB header:antitoxin/hydrolase
Chain: C: PDB Molecule:hicb;
PDBTitle: toxin-antitoxin complex from streptococcus pneumoniae
|
|
|
| 73 | c3rfqC_ |
|
not modelled |
32.8 |
23 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
|
|
| 74 | c3pvzD_ |
|
not modelled |
32.7 |
23 |
PDB header:lyase
Chain: D: PDB Molecule:udp-n-acetylglucosamine 4,6-dehydratase;
PDBTitle: udp-n-acetylglucosamine 4,6-dehydratase from vibrio fischeri
|
|
|
| 75 | c4w4tA_ |
|
not modelled |
31.7 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mxaa;
PDBTitle: the crystal structure of the terminal r domain from the myxalamid pks-2 nrps biosynthetic pathway
|
|
|
| 76 | d1vpxa_ |
|
not modelled |
30.7 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 77 | c2buiC_ |
|
not modelled |
30.4 |
40 |
PDB header:synthase
Chain: C: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] synthase i;
PDBTitle: e.coli beta-ketoacyl (acyl carrier protein) synthase i in2 complex with octanoic acid, 120k
|
|
|
| 78 | c5xvsA_ |
|
not modelled |
29.8 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:gdp/udp-n,n'-diacetylbacillosamine 2-epimerase
PDBTitle: crystal structure of udp-glcnac 2-epimerase neuc complexed with udp
|
|
|
| 79 | d1uuya_ |
|
not modelled |
29.5 |
18 |
Fold:Molybdenum cofactor biosynthesis proteins
Superfamily:Molybdenum cofactor biosynthesis proteins
Family:MogA-like |
|
|
| 80 | d1ub0a_ |
|
not modelled |
29.5 |
11 |
Fold:Ribokinase-like
Superfamily:Ribokinase-like
Family:Thiamin biosynthesis kinases |
|
|
| 81 | c4qqrB_ |
|
not modelled |
29.5 |
15 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3,5-epimerase/4-reductase;
PDBTitle: structural insight into nucleotide rhamnose synthase/epimerase-2 reductase from arabidopsis thaliana
|
|
|
| 82 | c3fryB_ |
|
not modelled |
28.5 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable copper-exporting p-type atpase a;
PDBTitle: crystal structure of the copa c-terminal metal binding domain
|
|
|
| 83 | c2el7A_ |
|
not modelled |
28.3 |
40 |
PDB header:ligase
Chain: A: PDB Molecule:tryptophanyl-trna synthetase;
PDBTitle: crystal structure of tryptophanyl-trna synthetase from thermus2 thermophilus
|
|
|
| 84 | c4pr3A_ |
|
not modelled |
28.1 |
35 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine nucleosidase / s-
PDBTitle: crystal structure of brucella melitensis 5'-methylthioadenosine/s-2 adenosylhomocysteine nucleosidase
|
|
|
| 85 | c3bl6A_ |
|
not modelled |
28.0 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine nucleosidase/s-
PDBTitle: crystal structure of staphylococcus aureus 5'-2 methylthioadenosine/s-adenosylhomocysteine nucleosidase in3 complex with formycin a
|
|
|
| 86 | d1j3na1 |
|
not modelled |
27.4 |
60 |
Fold:Thiolase-like
Superfamily:Thiolase-like
Family:Thiolase-related |
|
|
| 87 | c2h8gA_ |
|
not modelled |
27.3 |
12 |
PDB header:hydrolase
Chain: A: PDB Molecule:5'-methylthioadenosine nucleosidase;
PDBTitle: 5'-methylthioadenosine nucleosidase from arabidopsis2 thaliana
|
|
|
| 88 | c2a5hC_ |
|
not modelled |
26.6 |
10 |
PDB header:isomerase
Chain: C: PDB Molecule:l-lysine 2,3-aminomutase;
PDBTitle: 2.1 angstrom x-ray crystal structure of lysine-2,3-aminomutase from2 clostridium subterminale sb4, with michaelis analog (l-alpha-lysine3 external aldimine form of pyridoxal-5'-phosphate).
|
|
|
| 89 | d1fjha_ |
|
not modelled |
26.5 |
38 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 90 | d1iira_ |
|
not modelled |
26.3 |
26 |
Fold:UDP-Glycosyltransferase/glycogen phosphorylase
Superfamily:UDP-Glycosyltransferase/glycogen phosphorylase
Family:Gtf glycosyltransferase |
|
|
| 91 | c2kkhA_ |
|
not modelled |
26.1 |
19 |
PDB header:metal transport
Chain: A: PDB Molecule:putative heavy metal transporter;
PDBTitle: structure of the zinc binding domain of the atpase hma4
|
|
|
| 92 | c2qx7A_ |
|
not modelled |
25.3 |
63 |
PDB header:plant protein
Chain: A: PDB Molecule:eugenol synthase 1;
PDBTitle: structure of eugenol synthase from ocimum basilicum
|
|
|
| 93 | c4p7dA_ |
|
not modelled |
25.2 |
11 |
PDB header:toxin
Chain: A: PDB Molecule:antitoxin hicb3;
PDBTitle: antitoxin hicb3 crystal structure
|
|
|
| 94 | d1udca_ |
|
not modelled |
24.8 |
71 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 95 | c6aytD_ |
|
not modelled |
24.1 |
9 |
PDB header:hydrolase, transferase
Chain: D: PDB Molecule:5'-methylthioadenosine/s-adenosylhomocysteine nucleosidase;
PDBTitle: crystal structure of campylobacter jejuni 5'-methylthioadenosine/s-2 adenosyl homocysteine nucleosidase (mtan) complexed with3 pyrazinylthio-dadme-immucillin-a
|
|
|
| 96 | d1fmta2 |
|
not modelled |
24.0 |
17 |
Fold:Formyltransferase
Superfamily:Formyltransferase
Family:Formyltransferase |
|
|
| 97 | c3l77A_ |
|
not modelled |
23.8 |
38 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short-chain alcohol dehydrogenase;
PDBTitle: x-ray structure alcohol dehydrogenase from archaeon thermococcus2 sibiricus complexed with 5-hydroxy-nadp
|
|
|
| 98 | d1coza_ |
|
not modelled |
23.7 |
38 |
Fold:Adenine nucleotide alpha hydrolase-like
Superfamily:Nucleotidylyl transferase
Family:Cytidylyltransferase |
|
|
| 99 | d1r6da_ |
|
not modelled |
23.5 |
57 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 100 | c3icpA_ |
|
not modelled |
23.5 |
43 |
PDB header:isomerase
Chain: A: PDB Molecule:nad-dependent epimerase/dehydratase;
PDBTitle: crystal structure of udp-galactose 4-epimerase
|
|
|
| 101 | d2blla1 |
|
not modelled |
23.4 |
43 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 102 | c5mx6C_ |
|
not modelled |
23.4 |
7 |
PDB header:transferase
Chain: C: PDB Molecule:purine nucleoside phosphorylase deod-type;
PDBTitle: crystal structure of h. pylori purine nucleoside phosphorylase from2 clinical isolate hppnp-2
|
|
|
| 103 | c5intB_ |
|
not modelled |
23.2 |
25 |
PDB header:ligase
Chain: B: PDB Molecule:phosphopantothenate--cysteine ligase;
PDBTitle: crystal structure of the c-terminal domain of coenzyme a biosynthesis2 bifunctional protein coabc
|
|
|
| 104 | d2q46a1 |
|
not modelled |
23.0 |
36 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 105 | c3bsfB_ |
|
not modelled |
23.0 |
27 |
PDB header:hydrolase
Chain: B: PDB Molecule:at4g34840;
PDBTitle: crystal structure of the mta/sah nucleosidase
|
|
|
| 106 | c4zrmB_ |
|
not modelled |
22.9 |
57 |
PDB header:isomerase
Chain: B: PDB Molecule:udp-glucose 4-epimerase;
PDBTitle: crystal structure of udp-glucose 4-epimerase (tm0509) from2 hyperthermophilic eubacterium thermotoga maritima
|
|
|
| 107 | c1z34A_ |
|
not modelled |
22.7 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:purine nucleoside phosphorylase;
PDBTitle: crystal structure of trichomonas vaginalis purine nucleoside2 phosphorylase complexed with 2-fluoro-2'-deoxyadenosine
|
|
|
| 108 | c3lrfA_ |
|
not modelled |
22.6 |
60 |
PDB header:transferase
Chain: A: PDB Molecule:beta-ketoacyl synthase;
PDBTitle: crystal structure of beta-ketoacyl synthase from brucella2 melitensis
|
|
|
| 109 | c3s2uA_ |
|
not modelled |
22.5 |
32 |
PDB header:transferase
Chain: A: PDB Molecule:udp-n-acetylglucosamine--n-acetylmuramyl-(pentapeptide)
PDBTitle: crystal structure of the pseudomonas aeruginosa murg:udp-glcnac2 substrate complex
|
|
|
| 110 | c2pk3B_ |
|
not modelled |
22.4 |
43 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:gdp-6-deoxy-d-lyxo-4-hexulose reductase;
PDBTitle: crystal structure of a gdp-4-keto-6-deoxy-d-mannose reductase
|
|
|
| 111 | c4pvcB_ |
|
not modelled |
22.4 |
43 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph-dependent methylglyoxal reductase gre2;
PDBTitle: crystal structure of yeast methylglyoxal/ isovaleraldehyde reductase2 gre2
|
|
|
| 112 | c6bwlA_ |
|
not modelled |
22.1 |
71 |
PDB header:lyase
Chain: A: PDB Molecule:pal;
PDBTitle: x-ray structure of pal from bacillus thuringiensis
|
|
|
| 113 | d1gy8a_ |
|
not modelled |
22.0 |
57 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 114 | c1k82D_ |
|
not modelled |
21.8 |
11 |
PDB header:hydrolase/dna
Chain: D: PDB Molecule:formamidopyrimidine-dna glycosylase;
PDBTitle: crystal structure of e.coli formamidopyrimidine-dna2 glycosylase (fpg) covalently trapped with dna
|
|
|
| 115 | c3m2pD_ |
|
not modelled |
21.8 |
57 |
PDB header:isomerase
Chain: D: PDB Molecule:udp-n-acetylglucosamine 4-epimerase;
PDBTitle: the crystal structure of udp-n-acetylglucosamine 4-epimerase2 from bacillus cereus
|
|
|
| 116 | d1d8wa_ |
|
not modelled |
21.5 |
32 |
Fold:TIM beta/alpha-barrel
Superfamily:Xylose isomerase-like
Family:L-rhamnose isomerase |
|
|
| 117 | d1i24a_ |
|
not modelled |
21.5 |
28 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 118 | c2pzlB_ |
|
not modelled |
21.4 |
43 |
PDB header:sugar binding protein
Chain: B: PDB Molecule:putative nucleotide sugar epimerase/ dehydratase;
PDBTitle: crystal structure of the bordetella bronchiseptica enzyme wbmg in2 complex with nad and udp
|
|
|
| 119 | c2x4gA_ |
|
not modelled |
21.3 |
43 |
PDB header:isomerase
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: crystal structure of pa4631, a nucleoside-diphosphate-sugar epimerase2 from pseudomonas aeruginosa
|
|
|
| 120 | c4yl5A_ |
|
not modelled |
21.1 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:putative phosphomethylpyrimidine kinase;
PDBTitle: structure of a putative phosphomethylpyrimidine kinase from2 acinetobacter baumannii
|
|
|