1 c2qxxA_
100.0
99
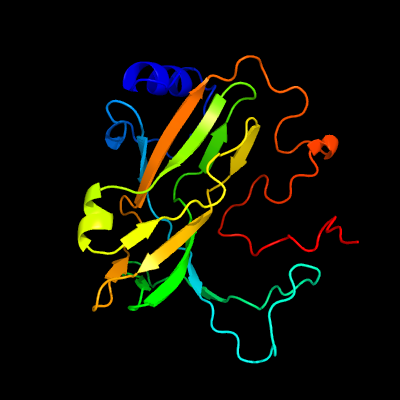
PDB header: hydrolaseChain: A: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: bifunctional dctp deaminase: dutpase from mycobacterium tuberculosis2 in complex with dttp
2 d1xs1a_
100.0
43
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
3 c4xjcD_
100.0
37
PDB header: hydrolaseChain: D: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: dctp deaminase-dutpase from bacillus halodurans
4 c2qlpC_
100.0
99
PDB header: hydrolaseChain: C: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: bifunctional dctp deaminase:dutpase from mycobacterium tuberculosis,2 apo form
5 c2r9qD_
100.0
25
PDB header: hydrolaseChain: D: PDB Molecule: 2'-deoxycytidine 5'-triphosphate deaminase;PDBTitle: crystal structure of 2'-deoxycytidine 5'-triphosphate deaminase from2 agrobacterium tumefaciens
6 d1pkha_
100.0
35
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
7 c2yzjB_
100.0
25
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: 167aa long hypothetical dutpase;PDBTitle: crystal structure of dctp deaminase from sulfolobus tokodaii
8 c3km3B_
100.0
28
PDB header: hydrolaseChain: B: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: crystal structure of eoxycytidine triphosphate deaminase from2 anaplasma phagocytophilum at 2.1a resolution
9 c4dhkB_
100.0
29
PDB header: hydrolaseChain: B: PDB Molecule: deoxycytidine triphosphate deaminase;PDBTitle: crystal structure of a deoxycytidine triphosphate deaminase (dctp2 deaminase) from burkholderia thailandensis
10 d1duna_
99.9
28
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
11 c3mbqC_
99.9
24
PDB header: hydrolaseChain: C: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of deoxyuridine 5-triphosphate nucleotidohydrolase2 from brucella melitensis, orthorhombic crystal form
12 d1f7ra_
99.9
32
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
13 c3ehwA_
99.9
24
PDB header: hydrolaseChain: A: PDB Molecule: dutp pyrophosphatase;PDBTitle: human dutpase in complex with alpha,beta-imido-dutp and mg2+:2 visualization of the full-length c-termini in all monomers and3 suggestion for an additional metal ion binding site
14 d1sixa_
99.9
29
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
15 c6maiA_
99.9
27
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of deoxyuridine 5'-triphosphate nucleotidohydrolase2 from legionella pneumophila philadelphia 1
16 c3tqzA_
99.9
32
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: structure of a deoxyuridine 5'-triphosphate nucleotidohydrolase (dut)2 from coxiella burnetii
17 c3so2A_
99.9
21
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: chlorella dutpase
18 c3ca9A_
99.9
25
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine triphosphatase;PDBTitle: evolution of chlorella virus dutpase
19 d1euwa_
99.9
24
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
20 d1rnja_
99.9
26
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
21 c3c3iA_
not modelled
99.9
25
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine triphosphatase;PDBTitle: evolution of chlorella virus dutpase
22 c3f4fB_
not modelled
99.9
22
PDB header: hydrolaseChain: B: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of dut1p, a dutpase from saccharomyces cerevisiae
23 c2okdB_
not modelled
99.9
27
PDB header: hydrolaseChain: B: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: high resolution crystal structures of vaccinia virus dutpase
24 d3ehwa1
not modelled
99.9
23
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
25 d1sjna_
not modelled
99.9
31
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
26 d1f7da_
not modelled
99.9
34
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
27 d1q5uz_
not modelled
99.9
25
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
28 c3lqwA_
not modelled
99.9
31
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: crystal structure of deoxyuridine 5-triphosphate2 nucleotidohydrolase from entamoeba histolytica
29 d1vyqa1
not modelled
99.9
23
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
30 c3h6xA_
not modelled
99.9
23
PDB header: hydrolaseChain: A: PDB Molecule: dutpase;PDBTitle: crystal structure of dutpase from streptococcus mutans
31 c2p9oB_
not modelled
99.9
21
PDB header: hydrolaseChain: B: PDB Molecule: dutp pyrophosphatase-like protein;PDBTitle: structure of dutpase from arabidopsis thaliana
32 c2bazA_
not modelled
99.9
21
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein bsu20020;PDBTitle: structure of yoss, a putative dutpase from bacillus subtilis
33 c5vjyC_
not modelled
99.9
21
PDB header: hydrolaseChain: C: PDB Molecule: dutp pyrophosphatase;PDBTitle: crystal structure of dutp pyrophosphatase protein, from naegleria2 fowleri
34 c3zf6A_
not modelled
99.9
28
PDB header: hydrolaseChain: A: PDB Molecule: dutpase;PDBTitle: phage dutpases control transfer of virulence genes by a proto-2 oncogenic g protein-like mechanism. (staphylococcus bacteriophage3 80alpha dutpase d81a d110c s168c mutant with dupnhpp).
35 c2d4nA_
not modelled
99.9
30
PDB header: hydrolaseChain: A: PDB Molecule: du;PDBTitle: crystal structure of m-pmv dutpase complexed with dupnpp, substrate2 analogue
36 c5y5pB_
not modelled
99.9
26
PDB header: viral proteinChain: B: PDB Molecule: wsv112;PDBTitle: crystal structure of the dutpase of white spot syndrome virus in2 complex with du,ppi and mg2+
37 c3ecyA_
not modelled
99.8
20
PDB header: hydrolaseChain: A: PDB Molecule: cg4584-pa, isoform a (bcdna.ld08534);PDBTitle: crystal structural analysis of drosophila melanogaster dutpase
38 d2bsya2
not modelled
99.8
24
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
39 d2bsya1
not modelled
99.8
26
Fold: beta-clipSuperfamily: dUTPase-likeFamily: dUTPase-like
40 c2bt1A_
not modelled
99.8
24
PDB header: hydrolaseChain: A: PDB Molecule: deoxyuridine 5'-triphosphate nucleotidohydrolase;PDBTitle: epstein barr virus dutpase in complex with a,b-imino dutp
41 c3p43A_
not modelled
28.4
19
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structure and activities of archaeal members of the ligd 3'2 phosphoesterase dna repair enzyme superfamily
42 d1tula_
not modelled
25.6
15
Fold: beta-clipSuperfamily: Tlp20, baculovirus telokin-like proteinFamily: Tlp20, baculovirus telokin-like protein
43 d1vioa2
not modelled
25.2
14
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Pseudouridine synthase RsuA N-terminal domain
44 c5dmpA_
not modelled
21.4
19
PDB header: hydrolaseChain: A: PDB Molecule: uncharacterized protein;PDBTitle: structure of the archaeal nhej phosphoesterase from methanocella2 paludicola.
45 c3n9dA_
not modelled
20.7
26
PDB header: ligaseChain: A: PDB Molecule: probable atp-dependent dna ligase;PDBTitle: monoclinic structure of p. aeruginosa ligd phosphoesterase domain
46 c1dm9A_
not modelled
20.1
13
PDB header: structural genomicsChain: A: PDB Molecule: hypothetical 15.5 kd protein in mrca-pcka intergenicPDBTitle: heat shock protein 15 kd
47 d1dm9a_
not modelled
20.1
13
Fold: Alpha-L RNA-binding motifSuperfamily: Alpha-L RNA-binding motifFamily: Heat shock protein 15 kD
48 c4mqdB_
not modelled
17.0
13
PDB header: hydrolase inhibitorChain: B: PDB Molecule: dna-entry nuclease inhibitor;PDBTitle: crystal structure of comj, inhibitor of the dna degrading activity of2 nuca, from bacillus subtilis
49 d1vqop1
not modelled
14.6
17
Fold: Ribosomal protein L19 (L19e)Superfamily: Ribosomal protein L19 (L19e)Family: Ribosomal protein L19 (L19e)
50 c1kskA_
not modelled
13.8
17
PDB header: lyaseChain: A: PDB Molecule: ribosomal small subunit pseudouridine synthase a;PDBTitle: structure of rsua
51 c3j21Q_
not modelled
13.7
21
PDB header: ribosomeChain: Q: PDB Molecule: 50s ribosomal protein l19e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
52 c2istA_
not modelled
13.4
10
PDB header: isomeraseChain: A: PDB Molecule: ribosomal large subunit pseudouridine synthase d;PDBTitle: crystal structure of rlud from e. coli
53 c4a17O_
not modelled
12.3
22
PDB header: ribosomeChain: O: PDB Molecule: rpl19;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 2.
54 d2visc_
not modelled
12.1
19
Fold: Viral protein domainSuperfamily: Viral protein domainFamily: Influenza hemagglutinin headpiece
55 c3iz5T_
not modelled
11.3
22
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l19 (l19e);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
56 c4a1eO_
not modelled
11.0
22
PDB header: ribosomeChain: O: PDB Molecule: rpl19;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna, 5.8s rrna3 and proteins of molecule 1
57 c4b6aR_
not modelled
10.9
17
PDB header: ribosomeChain: R: PDB Molecule: 60s ribosomal protein l19-b;PDBTitle: cryo-em structure of the 60s ribosomal subunit in complex2 with arx1 and rei1
58 d2viua_
not modelled
10.7
19
Fold: Viral protein domainSuperfamily: Viral protein domainFamily: Influenza hemagglutinin headpiece
59 c4a1cO_
not modelled
10.3
21
PDB header: ribosomeChain: O: PDB Molecule: rpl19;PDBTitle: t.thermophila 60s ribosomal subunit in complex with2 initiation factor 6. this file contains 5s rrna,3 5.8s rrna and proteins of molecule 4.
60 c3p4hA_
not modelled
10.2
14
PDB header: hydrolaseChain: A: PDB Molecule: atp-dependent dna ligase, n-terminal domain protein;PDBTitle: structures of archaeal members of the ligd 3'-phosphoesterase dna2 repair enzyme superfamily
61 c3j39R_
not modelled
10.1
26
PDB header: ribosomeChain: R: PDB Molecule: 60s ribosomal protein l19;PDBTitle: structure of the d. melanogaster 60s ribosomal proteins
62 d1mqma_
not modelled
9.9
19
Fold: Viral protein domainSuperfamily: Viral protein domainFamily: Influenza hemagglutinin headpiece
63 c5hmqE_
not modelled
9.8
31
PDB header: lyaseChain: E: PDB Molecule: 4-hydroxyphenylpyruvate dioxygenase;PDBTitle: xylose isomerase-like tim barrel/4-hydroxyphenylpyruvate dioxygenase2 fusion protein
64 c3zf7T_
not modelled
9.7
28
PDB header: ribosomeChain: T: PDB Molecule: 60s ribosomal protein l19, putative;PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
65 c1ha0A_
not modelled
9.0
19
PDB header: viral proteinChain: A: PDB Molecule: protein (hemagglutinin precursor);PDBTitle: hemagglutinin precursor ha0
66 c5z81A_
not modelled
8.9
11
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 15;PDBTitle: trimeric structure of vibrio cholerae heat shock protein 15 at 2.32 angstrom resolution
67 c2ahqA_
not modelled
8.6
12
PDB header: transcriptionChain: A: PDB Molecule: rna polymerase sigma factor rpon;PDBTitle: solution structure of the c-terminal rpon domain of sigma-2 54 from aquifex aeolicus
68 c4uj3B_
not modelled
8.6
12
PDB header: transport proteinChain: B: PDB Molecule: rab-3a-interacting protein;PDBTitle: crystal structure of human rab11-rabin8-fip3
69 c3mpbA_
not modelled
8.3
17
PDB header: isomeraseChain: A: PDB Molecule: sugar isomerase;PDBTitle: z5688 from e. coli o157:h7 bound to fructose
70 c5oy0m_
not modelled
7.0
33
PDB header: photosynthesisChain: M: PDB Molecule: photosystem i reaction center subunit xii;PDBTitle: structure of synechocystis photosystem i trimer at 2.5a resolution
71 c5oy09_
not modelled
7.0
33
PDB header: photosynthesisChain: 9: PDB Molecule: photosystem i reaction center subunit xii;PDBTitle: structure of synechocystis photosystem i trimer at 2.5a resolution
72 c6hqbM_
not modelled
7.0
33
PDB header: photosynthesisChain: M: PDB Molecule: photosystem i reaction center subunit xii;PDBTitle: monomeric cyanobacterial photosystem i
73 c5oy0M_
not modelled
7.0
33
PDB header: photosynthesisChain: M: PDB Molecule: photosystem i reaction center subunit xii;PDBTitle: structure of synechocystis photosystem i trimer at 2.5a resolution
74 c4l6vM_
not modelled
7.0
33
PDB header: electron transportChain: M: PDB Molecule: photosystem i reaction center subunit xii;PDBTitle: crystal structure of a virus like photosystem i from the2 cyanobacterium synechocystis pcc 6803
75 c4l6v7_
not modelled
7.0
33
PDB header: electron transportChain: 7: PDB Molecule: photosystem i reaction center subunit xii;PDBTitle: crystal structure of a virus like photosystem i from the2 cyanobacterium synechocystis pcc 6803
76 d1v54b1
not modelled
6.9
27
Fold: Cupredoxin-likeSuperfamily: CupredoxinsFamily: Periplasmic domain of cytochrome c oxidase subunit II
77 c2zkrp_
not modelled
6.7
28
PDB header: ribosomal protein/rnaChain: P: PDB Molecule: rna expansion segment es31 part i;PDBTitle: structure of a mammalian ribosomal 60s subunit within an 80s complex2 obtained by docking homology models of the rna and proteins into an3 8.7 a cryo-em map
78 c2wwaJ_
not modelled
6.6
20
PDB header: ribosomeChain: J: PDB Molecule: 60s ribosomal protein l19;PDBTitle: cryo-em structure of idle yeast ssh1 complex bound to the yeast 80s2 ribosome
79 c5dudB_
not modelled
6.5
36
PDB header: unknown functionChain: B: PDB Molecule: ybgj;PDBTitle: crystal structure of e. coli ybgjk
80 c5olpB_
not modelled
6.4
20
PDB header: hydrolaseChain: B: PDB Molecule: pectate lyase;PDBTitle: galacturonidase
81 c2psbA_
not modelled
6.4
18
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: yerb protein;PDBTitle: crystal structure of yerb protein from bacillus subtilis. northeast2 structural genomics target sr586
82 d2psba1
not modelled
6.4
18
Fold: YerB-likeSuperfamily: YerB-likeFamily: YerB-like
83 d2q02a1
not modelled
5.8
21
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
84 d1g6ea_
not modelled
5.5
28
Fold: gamma-Crystallin-likeSuperfamily: gamma-Crystallin-likeFamily: Antifungal protein AFP1
85 d1uwfa1
not modelled
5.5
19
Fold: Common fold of diphtheria toxin/transcription factors/cytochrome fSuperfamily: Bacterial adhesinsFamily: Pilus subunits
86 d1zx5a1
not modelled
5.3
17
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: Type I phosphomannose isomerase