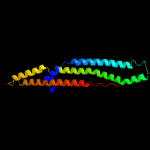
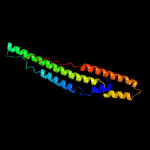
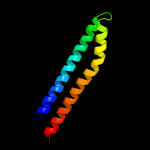
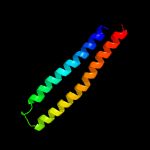
1 c5xfsB_
100.0
55
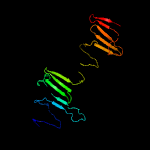
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
2 d2g38b1
100.0
34
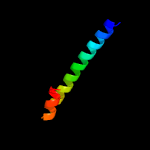
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
3 c2g38B_
100.0
34
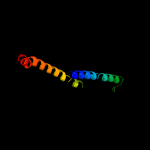
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
4 c4xy3A_
100.0
18
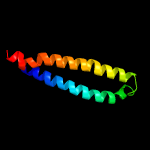
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion-associated protein espb;PDBTitle: structure of esx-1 secreted protein espb
5 c2vs0B_
96.8
10
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
6 c4iogD_
96.8
17
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
7 c4wj2A_
96.7
18
PDB header: unknown functionChain: A: PDB Molecule: antigen mtb48;PDBTitle: mycobacterial protein
8 c3gvmA_
96.4
10
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 c3zbhC_
96.1
16
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
10 d1wa8a1
95.0
20
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
11 c5nxhB_
91.1
12
PDB header: viral proteinChain: B: PDB Molecule: long-tail fiber proximal subunit;PDBTitle: crystal structure of the carboxy-terminal region of the bacteriophage2 t4 proximal long tail fibre protein gp34, residues 744-1289 at 2.93 angstrom resolution
12 c3jywF_
88.1
46
PDB header: ribosomeChain: F: PDB Molecule: 60s ribosomal protein l7(a);PDBTitle: structure of the 60s proteins for eukaryotic ribosome based on cryo-em2 map of thermomyces lanuginosus ribosome at 8.9a resolution
13 c4i0xA_
84.5
21
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
14 c2vsnB_
82.8
21
PDB header: transferaseChain: B: PDB Molecule: xcogt;PDBTitle: structure and topological arrangement of an o-glcnac2 transferase homolog: insight into molecular control of3 intracellular glycosylation
15 c4lwsA_
80.9
19
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
16 d1wa8b1
80.1
21
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
17 d1ui5a2
70.9
18
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
18 d1rp4a_
64.6
16
Fold: ERO1-likeSuperfamily: ERO1-likeFamily: ERO1-like
19 c4lwsB_
61.4
13
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
20 c3pe3D_
60.4
18
PDB header: transferaseChain: D: PDB Molecule: udp-n-acetylglucosamine--peptide n-PDBTitle: structure of human o-glcnac transferase and its complex with a peptide2 substrate
21 c3lhoA_
not modelled
60.1
44
PDB header: hydrolaseChain: A: PDB Molecule: putative hydrolase;PDBTitle: crystal structure of putative hydrolase (yp_751971.1) from shewanella2 frigidimarina ncimb 400 at 1.80 a resolution
22 c5djsA_
not modelled
58.2
19
PDB header: transferaseChain: A: PDB Molecule: tetratricopeptide tpr_2 repeat protein;PDBTitle: thermobaculum terrenum o-glcnac transferase mutant - k341m
23 c2kg7B_
not modelled
57.5
19
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
24 c3ahrA_
not modelled
56.4
19
PDB header: oxidoreductaseChain: A: PDB Molecule: ero1-like protein alpha;PDBTitle: inactive human ero1
25 c4rqoB_
not modelled
54.7
18
PDB header: lyaseChain: B: PDB Molecule: l-serine dehydratase;PDBTitle: crystal structure of l-serine dehydratase from legionella pneumophila
26 d3cx5d1
not modelled
54.4
40
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain
27 d1ppjd1
not modelled
53.1
35
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Cytochrome bc1 domain
28 c5azaA_
not modelled
52.8
19
PDB header: sugar binding protein, transferaseChain: A: PDB Molecule: maltose-binding periplasmic protein,oligosaccharylPDBTitle: crystal structure of mbp-saglb fusion protein with a 20-residue spacer2 in the connector helix
29 c4f9iA_
not modelled
52.7
15
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase/delta-1-pyrroline-5-carboxylatePDBTitle: crystal structure of proline utilization a (puta) from geobacter2 sulfurreducens pca
30 c4jzaB_
not modelled
51.4
41
PDB header: hydrolaseChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a legionella phosphoinositide phosphatase:2 insights into lipid metabolism in pathogen host interaction
31 c1zrtD_
not modelled
46.3
50
PDB header: oxidoreductase/metal transportChain: D: PDB Molecule: cytochrome c1;PDBTitle: rhodobacter capsulatus cytochrome bc1 complex with2 stigmatellin bound
32 c6ghrF_
not modelled
46.2
59
PDB header: photosynthesisChain: F: PDB Molecule: cp12 polypeptide;PDBTitle: cyanobacterial gapdh with full-length cp12
33 d1exta3
not modelled
45.2
57
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
34 d1ncfb3
not modelled
45.1
57
Fold: TNF receptor-likeSuperfamily: TNF receptor-likeFamily: TNF receptor-like
35 c6fhjA_
not modelled
44.7
18
PDB header: hydrolaseChain: A: PDB Molecule: protein,protein;PDBTitle: structural dynamics and catalytic properties of a multi-modular2 xanthanase, native.
36 c3fpjA_
not modelled
44.3
17
PDB header: biosynthetic protein, transferaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of e81q mutant of mtnas in complex with s-2 adenosylmethionine
37 c3cwbQ_
not modelled
44.1
35
PDB header: oxidoreductaseChain: Q: PDB Molecule: mitochondrial cytochrome c1, heme protein;PDBTitle: chicken cytochrome bc1 complex inhibited by an iodinated analogue of2 the polyketide crocacin-d
38 c1qcrD_
not modelled
43.2
35
PDB header: oxidoreductaseChain: D: PDB Molecule: ubiquinol cytochrome c oxidoreductase;PDBTitle: crystal structure of bovine mitochondrial cytochrome bc12 complex, alpha carbon atoms only
39 c3b5oA_
not modelled
42.9
50
PDB header: oxidoreductaseChain: A: PDB Molecule: cadd-like protein of unknown function;PDBTitle: crystal structure of a cadd-like protein of unknown function2 (npun_f6505) from nostoc punctiforme pcc 73102 at 1.35 a resolution
40 d2a15a1
not modelled
42.7
21
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: Ketosteroid isomerase-like
41 c3qx3B_
not modelled
42.4
35
PDB header: isomerase/dna/isomerase inhibitorChain: B: PDB Molecule: dna topoisomerase 2-beta;PDBTitle: human topoisomerase iibeta in complex with dna and etoposide
42 c4nl6C_
not modelled
42.4
71
PDB header: splicingChain: C: PDB Molecule: survival motor neuron protein;PDBTitle: structure of the full-length form of the protein smn found in healthy2 patients
43 c4yfbD_
not modelled
42.3
24
PDB header: hydrolaseChain: D: PDB Molecule: protein related to penicillin acylase;PDBTitle: structure of n-acylhomoserine lactone acylase macq in complex with2 phenylacetic acid
44 c6fhnA_
not modelled
42.2
18
PDB header: hydrolaseChain: A: PDB Molecule: protein;PDBTitle: structural dynamics and catalytic properties of a multi-modular2 xanthanase (pt derivative)
45 c4fm9A_
not modelled
42.0
30
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase 2-alpha;PDBTitle: human topoisomerase ii alpha bound to dna
46 d1xkna_
not modelled
41.7
14
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
47 d2fr1a1
not modelled
40.6
20
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Tyrosine-dependent oxidoreductases
48 c4lrvL_
not modelled
40.5
40
PDB header: dna binding proteinChain: L: PDB Molecule: dna sulfur modification protein dnde;PDBTitle: crystal structure of dnde from escherichia coli b7a involved in dna2 phosphorothioation modification
49 d1byra_
not modelled
38.5
28
Fold: Phospholipase D/nucleaseSuperfamily: Phospholipase D/nucleaseFamily: Nuclease
50 c2yiuE_
not modelled
38.2
50
PDB header: oxidoreductaseChain: E: PDB Molecule: cytochrome c1, heme protein;PDBTitle: x-ray structure of the dimeric cytochrome bc1 complex from2 the soil bacterium paracoccus denitrificans at 2.73 angstrom resolution
51 d1de4c2
not modelled
37.7
3
Fold: The "swivelling" beta/beta/alpha domainSuperfamily: PA domainFamily: PA domain
52 c4urjA_
not modelled
37.3
13
PDB header: unknown functionChain: A: PDB Molecule: protein fam83a;PDBTitle: crystal structure of human bj-tsa-9
53 c4q7oA_
not modelled
37.1
38
PDB header: immune systemChain: A: PDB Molecule: immunity protein;PDBTitle: the crystal structure of an immunity protein nmb0503 from neisseria2 meningitidis mc58
54 d2gxfa1
not modelled
36.9
12
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: YybH-like
55 c1p84D_
not modelled
36.7
40
PDB header: oxidoreductaseChain: D: PDB Molecule: cytochrome c1, heme protein;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
56 d1luaa2
not modelled
36.4
18
Fold: Aminoacid dehydrogenase-like, N-terminal domainSuperfamily: Aminoacid dehydrogenase-like, N-terminal domainFamily: Methylene-tetrahydromethanopterin dehydrogenase
57 c5lzkB_
not modelled
36.3
4
PDB header: structural genomicsChain: B: PDB Molecule: protein fam83b;PDBTitle: structure of the domain of unknown function duf1669 from human fam83b
58 c5aizA_
not modelled
35.2
15
PDB header: zinc-binding proteinChain: A: PDB Molecule: zinc finger miz domain-containing protein 1;PDBTitle: the pias-like coactivator zmiz1 is a direct and selective cofactor2 of notch1 in t-cell development and leukemia
59 c3vohA_
not modelled
34.9
13
PDB header: hydrolaseChain: A: PDB Molecule: cellobiohydrolase;PDBTitle: cccel6a catalytic domain complexed with cellobiose
60 c2vakF_
not modelled
34.9
27
PDB header: viral proteinChain: F: PDB Molecule: sigma a;PDBTitle: crystal structure of the avian reovirus inner capsid protein sigmaa
61 c2pheC_
not modelled
34.7
31
PDB header: transcriptionChain: C: PDB Molecule: alpha trans-inducing protein;PDBTitle: model for vp16 binding to pc4
62 c2fynH_
not modelled
34.6
35
PDB header: oxidoreductaseChain: H: PDB Molecule: cytochrome c1;PDBTitle: crystal structure analysis of the double mutant rhodobacter2 sphaeroides bc1 complex
63 c2jg7G_
not modelled
34.0
16
PDB header: oxidoreductaseChain: G: PDB Molecule: antiquitin;PDBTitle: crystal structure of seabream antiquitin and elucidation of2 its substrate specificity
64 c2wybA_
not modelled
33.5
15
PDB header: hydrolaseChain: A: PDB Molecule: acyl-homoserine lactone acylase pvdq subunitPDBTitle: the quorum quenching n-acyl homoserine lactone acylase pvdq2 with a covalently bound dodecanoic acid
65 c3pefA_
not modelled
32.9
11
PDB header: oxidoreductaseChain: A: PDB Molecule: 6-phosphogluconate dehydrogenase, nad-binding;PDBTitle: crystal structure of gamma-hydroxybutyrate dehydrogenase from2 geobacter metallireducens in complex with nadp+
66 c2k2uB_
not modelled
32.7
31
PDB header: transcriptionChain: B: PDB Molecule: alpha trans-inducing protein;PDBTitle: nmr structure of the complex between tfb1 subunit of tfiih2 and the activation domain of vp16
67 c2wk1A_
not modelled
32.3
28
PDB header: transferaseChain: A: PDB Molecule: novp;PDBTitle: structure of the o-methyltransferase novp
68 d3cu3a1
not modelled
32.3
16
Fold: Cystatin-likeSuperfamily: NTF2-likeFamily: BaiE/LinA-like
69 c3rf7A_
not modelled
31.8
13
PDB header: oxidoreductaseChain: A: PDB Molecule: iron-containing alcohol dehydrogenase;PDBTitle: crystal structure of an iron-containing alcohol dehydrogenase2 (sden_2133) from shewanella denitrificans os-217 at 2.12 a resolution
70 c5kf6B_
not modelled
31.8
21
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional protein puta;PDBTitle: structure of proline utilization a from sinorhizobium meliloti2 complexed with l-tetrahydrofuroic acid and nad+ in space group p21
71 d1bjta_
not modelled
31.6
22
Fold: Type II DNA topoisomeraseSuperfamily: Type II DNA topoisomeraseFamily: Type II DNA topoisomerase
72 c1bjtA_
not modelled
31.6
22
PDB header: topoisomeraseChain: A: PDB Molecule: topoisomerase ii;PDBTitle: topoisomerase ii residues 409-1201
73 c3hx8A_
not modelled
30.9
23
PDB header: isomeraseChain: A: PDB Molecule: putative ketosteroid isomerase;PDBTitle: crystal structure of putative ketosteroid isomerase (np_103587.1) from2 mesorhizobium loti at 1.45 a resolution
74 c3s8mA_
not modelled
30.5
14
PDB header: oxidoreductaseChain: A: PDB Molecule: enoyl-acp reductase;PDBTitle: the crystal structure of fabv
75 d1joga_
not modelled
30.0
17
Fold: Four-helical up-and-down bundleSuperfamily: Nucleotidyltransferase substrate binding subunit/domainFamily: Family 1 bi-partite nucleotidyltransferase subunit
76 c6iz9B_
not modelled
29.9
15
PDB header: transferaseChain: B: PDB Molecule: beta-transaminase;PDBTitle: crystal structure of the apo form of a beta-transaminase from2 mesorhizobium sp. strain luk
77 c4gfhA_
not modelled
29.9
23
PDB header: isomerase/dnaChain: A: PDB Molecule: dna topoisomerase 2;PDBTitle: topoisomerase ii-dna-amppnp complex
78 d1jpdx2
not modelled
29.9
21
Fold: Enolase N-terminal domain-likeSuperfamily: Enolase N-terminal domain-likeFamily: Enolase N-terminal domain-like
79 c2ve5H_
not modelled
29.6
15
PDB header: oxidoreductaseChain: H: PDB Molecule: betaine aldehyde dehydrogenase;PDBTitle: crystallographic structure of betaine aldehyde2 dehydrogenase from pseudomonas aeruginosa
80 c3q3hA_
not modelled
29.0
13
PDB header: transferaseChain: A: PDB Molecule: hmw1c-like glycosyltransferase;PDBTitle: crystal structure of the actinobacillus pleuropneumoniae hmw1c2 glycosyltransferase in complex with udp-glc
81 d1vkpa_
not modelled
28.7
28
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
82 d1bvsa1
not modelled
28.0
39
Fold: RuvA C-terminal domain-likeSuperfamily: DNA helicase RuvA subunit, C-terminal domainFamily: DNA helicase RuvA subunit, C-terminal domain
83 d1oy0a_
not modelled
27.8
23
Fold: TIM beta/alpha-barrelSuperfamily: Phosphoenolpyruvate/pyruvate domainFamily: Ketopantoate hydroxymethyltransferase PanB
84 c3r8rJ_
not modelled
27.0
16
PDB header: transferaseChain: J: PDB Molecule: transaldolase;PDBTitle: transaldolase from bacillus subtilis
85 c6ajrA_
not modelled
27.0
27
PDB header: dna binding proteinChain: A: PDB Molecule: uracil dna glycosylase superfamily protein;PDBTitle: complex form of uracil dna glycosylase x and uracil
86 c2v7iA_
not modelled
27.0
29
PDB header: biosynthetic proteinChain: A: PDB Molecule: prnb;PDBTitle: prnb native
87 c3hazA_
not modelled
26.7
24
PDB header: oxidoreductaseChain: A: PDB Molecule: proline dehydrogenase;PDBTitle: crystal structure of bifunctional proline utilization a2 (puta) protein
88 c3ly8A_
not modelled
26.4
14
PDB header: signaling proteinChain: A: PDB Molecule: transcriptional activator cadc;PDBTitle: crystal structure of mutant d471e of the periplasmic domain of cadc
89 d2rh3a1
not modelled
26.1
24
Fold: Ribbon-helix-helixSuperfamily: Ribbon-helix-helixFamily: VirC2-like
90 c2phgB_
not modelled
26.0
31
PDB header: transcriptionChain: B: PDB Molecule: alpha trans-inducing protein;PDBTitle: model for vp16 binding to tfiib
91 d1vefa1
not modelled
25.9
18
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: GABA-aminotransferase-like
92 c4xvzB_
not modelled
25.7
28
PDB header: transferaseChain: B: PDB Molecule: mycinamicin iii 3''-o-methyltransferase;PDBTitle: mycf mycinamicin iii 3'-o-methyltransferase in complex with mg
93 c3sdoB_
not modelled
25.7
13
PDB header: oxidoreductaseChain: B: PDB Molecule: nitrilotriacetate monooxygenase;PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
94 c2ordA_
not modelled
25.6
16
PDB header: transferaseChain: A: PDB Molecule: acetylornithine aminotransferase;PDBTitle: crystal structure of acetylornithine aminotransferase (ec 2.6.1.11)2 (acoat) (tm1785) from thermotoga maritima at 1.40 a resolution
95 c4h7nA_
not modelled
25.5
16
PDB header: oxidoreductaseChain: A: PDB Molecule: aldehyde dehydrogenase;PDBTitle: the structure of putative aldehyde dehydrogenase puta from anabaena2 variabilis.
96 c6ca8A_
not modelled
25.4
23
PDB header: isomeraseChain: A: PDB Molecule: dna topoisomerase 2;PDBTitle: crystal structure of plasmodium falciparum topoisomerase ii dna-2 binding, cleavage and re-ligation domain
97 d1c9ka_
not modelled
25.3
21
Fold: P-loop containing nucleoside triphosphate hydrolasesSuperfamily: P-loop containing nucleoside triphosphate hydrolasesFamily: RecA protein-like (ATPase-domain)
98 c4i3wC_
not modelled
25.2
26
PDB header: oxidoreductaseChain: C: PDB Molecule: aldehyde dehydrogenase (nad+);PDBTitle: structure of phosphonoacetaldehyde dehydrogenase in complex with2 gylceraldehyde-3-phosphate and cofactor nad+
99 c3r5zB_
not modelled
25.2
22
PDB header: unknown functionChain: B: PDB Molecule: putative uncharacterized protein;PDBTitle: structure of a deazaflavin-dependent reductase from nocardia2 farcinica, with co-factor f420
100 c3u4jB_
not modelled
24.9
21
PDB header: oxidoreductaseChain: B: PDB Molecule: nad-dependent aldehyde dehydrogenase;PDBTitle: crystal structure of nad-dependent aldehyde dehydrogenase from2 sinorhizobium meliloti
101 c2axtc_
not modelled
24.8
35
PDB header: electron transportChain: C: PDB Molecule: photosystem ii cp43 protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
102 d2axtc1
not modelled
24.8
35
Fold: Photosystem II antenna protein-likeSuperfamily: Photosystem II antenna protein-likeFamily: Photosystem II antenna protein-like
103 d1tpla_
not modelled
24.8
16
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Beta-eliminating lyases
104 c6cboB_
not modelled
24.5
23
PDB header: transferaseChain: B: PDB Molecule: c-6' aminotransferase;PDBTitle: x-ray structure of genb1 from micromonospora echinospora in complex2 with neamine and plp (as the external aldimine)
105 d1vpxa_
not modelled
24.5
11
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
106 d2ewoa1
not modelled
24.4
28
Fold: Pentein, beta/alpha-propellerSuperfamily: PenteinFamily: Porphyromonas-type peptidylarginine deiminase
107 c3kb4D_
not modelled
24.3
19
PDB header: structural genomics, unknown functionChain: D: PDB Molecule: alr8543 protein;PDBTitle: crystal structure of the alr8543 protein in complex with2 geranylgeranyl monophosphate and magnesium ion from nostoc sp. pcc3 7120, northeast structural genomics consortium target nsr141
108 c5frgA_
not modelled
24.3
63
PDB header: protein bindingChain: A: PDB Molecule: formin-binding protein 1-like;PDBTitle: the nmr structure of the cdc42-interacting region of toca1
109 d1wx0a1
not modelled
24.0
13
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I aldolase
110 c2h7fX_
not modelled
24.0
18
PDB header: isomerase/dnaChain: X: PDB Molecule: dna topoisomerase 1;PDBTitle: structure of variola topoisomerase covalently bound to dna
111 c3k66A_
not modelled
23.7
26
PDB header: cell adhesionChain: A: PDB Molecule: beta-amyloid-like protein;PDBTitle: x-ray crystal structure of the e2 domain of c. elegans apl-1
112 c5mqrA_
not modelled
23.6
15
PDB header: hydrolaseChain: A: PDB Molecule: beta-l-arabinobiosidase;PDBTitle: sialidase bt_1020
113 c3fyqA_
not modelled
23.3
17
PDB header: cell adhesionChain: A: PDB Molecule: cg6831-pa (talin);PDBTitle: structure of drosophila melanogaster talin ibs2 domain (residues 1981-2 2168)
114 c5cgaC_
not modelled
23.3
19
PDB header: transferaseChain: C: PDB Molecule: hydroxyethylthiazole kinase;PDBTitle: structure of hydroxyethylthiazole kinase thim from staphylococcus2 aureus in complex with substrate analog 2-(1,3,5-trimethyl-1h-3 pyrazole-4-yl)ethanol
115 c4evxA_
not modelled
23.1
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative phage endolysin;PDBTitle: crystal structure of putative phage endolysin from s. enterica
116 d2f7fa1
not modelled
23.1
19
Fold: TIM beta/alpha-barrelSuperfamily: Nicotinate/Quinolinate PRTase C-terminal domain-likeFamily: NadC C-terminal domain-like
117 c2yikA_
not modelled
23.0
10
PDB header: hydrolaseChain: A: PDB Molecule: endoglucanase;PDBTitle: catalytic domain of clostridium thermocellum celt
118 c2nwqA_
not modelled
23.0
13
PDB header: oxidoreductaseChain: A: PDB Molecule: probable short-chain dehydrogenase;PDBTitle: short chain dehydrogenase from pseudomonas aeruginosa
119 c5jn6A_
not modelled
22.9
89
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: the nmr solution structure of rpa3313
120 d1yymg1
not modelled
22.9
33
Fold: gp120 coreSuperfamily: gp120 coreFamily: gp120 core