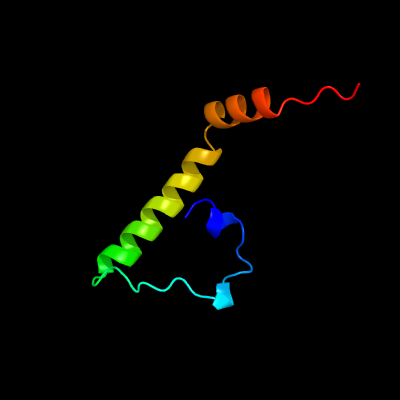
1 d1bxca_
76.6
32
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
2 c5v2gB_
61.2
50
PDB header: de novo proteinChain: B: PDB Molecule: 20-mer peptide;PDBTitle: de novo design of novel covalent constrained meso-size peptide2 scaffolds with unique tertiary structures
3 c5v2gA_
61.2
50
PDB header: de novo proteinChain: A: PDB Molecule: 20-mer peptide;PDBTitle: de novo design of novel covalent constrained meso-size peptide2 scaffolds with unique tertiary structures
4 c5v2gC_
61.2
50
PDB header: de novo proteinChain: C: PDB Molecule: 20-mer peptide;PDBTitle: de novo design of novel covalent constrained meso-size peptide2 scaffolds with unique tertiary structures
5 c5fjsB_
58.3
14
PDB header: hydrolaseChain: B: PDB Molecule: glucosylceramidase;PDBTitle: bacterial beta-glucosidase reveals the structural and functional2 basis of genetic defects in human glucocerebrosidase 2 (gba2)3 disorders
6 d1bxba_
55.7
31
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
7 c2ww1B_
53.8
19
PDB header: hydrolaseChain: B: PDB Molecule: putative alpha-1,2-mannosidase;PDBTitle: structure of the family gh92 inverting mannosidase bt39902 from bacteroides thetaiotaomicron vpi-5482 in complex with3 thiomannobioside
8 c5swiD_
37.3
16
PDB header: hydrolaseChain: D: PDB Molecule: sugar hydrolase;PDBTitle: crystal structure of spgh92 in complex with mannose
9 c3f1iS_
36.4
28
PDB header: protein bindingChain: S: PDB Molecule: signal transducing adapter molecule 1;PDBTitle: human escrt-0 core complex
10 c3ktcB_
30.6
11
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase;PDBTitle: crystal structure of putative sugar isomerase (yp_050048.1) from2 erwinia carotovora atroseptica scri1043 at 1.54 a resolution
11 c4ozwA_
30.4
23
PDB header: lyaseChain: A: PDB Molecule: alginate lyase;PDBTitle: crystal structure of the periplasmic alginate lyase algl h202a mutant
12 c2l3gA_
21.7
31
PDB header: signaling proteinChain: A: PDB Molecule: rho guanine nucleotide exchange factor 7;PDBTitle: solution nmr structure of ch domain of rho guanine nucleotide exchange2 factor 7 from homo sapiens, northeast structural genomics consortium3 target hr4495e
13 d1iq0a1
21.2
22
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
14 c3mkqB_
20.2
24
PDB header: transport proteinChain: B: PDB Molecule: coatomer subunit alpha;PDBTitle: crystal structure of yeast alpha/betaprime-cop subcomplex of the copi2 vesicular coat
15 c4madA_
17.8
23
PDB header: hydrolaseChain: A: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of beta-galactosidase c (bgac) from bacillus2 circulans atcc 31382
16 c2ihmA_
16.9
42
PDB header: transferase/dnaChain: A: PDB Molecule: dna polymerase mu;PDBTitle: polymerase mu in ternary complex with gapped 11mer dna duplex and2 bound incoming nucleotide
17 d1f7ua1
16.2
17
Fold: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesSuperfamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetasesFamily: Anticodon-binding domain of a subclass of class I aminoacyl-tRNA synthetases
18 c4c9bB_
15.6
19
PDB header: splicingChain: B: PDB Molecule: pre-mrna-splicing factor cwc22 homolog;PDBTitle: crystal structure of eif4aiii-cwc22 complex
19 c2m35A_
15.4
78
PDB header: toxinChain: A: PDB Molecule: k-ssm1a;PDBTitle: nmr study of k-ssm1a
20 d1muwa_
14.9
31
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
21 c4zlgA_
not modelled
14.8
14
PDB header: transferaseChain: A: PDB Molecule: putative b-glycan phosphorylase;PDBTitle: cellobionic acid phosphorylase - gluconic acid complex
22 d1c6vx_
not modelled
13.6
71
Fold: SH3-like barrelSuperfamily: DNA-binding domain of retroviral integraseFamily: DNA-binding domain of retroviral integrase
23 c1c6vX_
not modelled
13.6
71
PDB header: dna binding proteinChain: X: PDB Molecule: protein (siu89134);PDBTitle: siv integrase (catalytic domain + dna biding domain comprising2 residues 50-293) mutant with phe 185 replaced by his (f185h)
24 d1qt1a_
not modelled
12.9
31
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
25 c4gmnA_
not modelled
12.1
13
PDB header: rna binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structural basis of rpl5 recognition by syo1
26 c2gm4B_
not modelled
12.0
30
PDB header: recombination, dnaChain: B: PDB Molecule: transposon gamma-delta resolvase;PDBTitle: an activated, tetrameric gamma-delta resolvase: hin chimaera bound to2 cleaved dna
27 c3ty8A_
not modelled
11.6
32
PDB header: transferaseChain: A: PDB Molecule: polynucleotide 2',3'-cyclic phosphate phosphodiesterase /PDBTitle: crystal structure of c. thermocellum pnkp ligase domain apo form
28 c4e6nC_
not modelled
11.4
32
PDB header: protein bindingChain: C: PDB Molecule: metallophosphoesterase;PDBTitle: crystal structure of bacterial pnkp-c/hen1-n heterodimer
29 c1wynA_
not modelled
11.1
24
PDB header: structural proteinChain: A: PDB Molecule: calponin-2;PDBTitle: solution structure of the ch domain of human calponin-2
30 c5wftA_
not modelled
9.9
20
PDB header: structural proteinChain: A: PDB Molecule: pelb;PDBTitle: pelb 319-436 from pseudomonas aeruginosa pao1
31 c4luqD_
not modelled
9.3
45
PDB header: protein binding/toxin inhibitorChain: D: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of virulence effector tse3 in complex with2 neutralizer tsi3
32 d2pqrb1
not modelled
9.3
21
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)
33 c5a1uC_
not modelled
9.2
28
PDB header: transport proteinChain: C: PDB Molecule: coatomer subunit alpha;PDBTitle: the structure of the copi coat triad
34 d1y8ma1
not modelled
9.2
21
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)
35 c5dseC_
not modelled
9.1
17
PDB header: protein bindingChain: C: PDB Molecule: tetratricopeptide repeat protein 7b;PDBTitle: crystal structure of the ttc7b/hyccin complex
36 c3p6lA_
not modelled
9.1
19
PDB header: isomeraseChain: A: PDB Molecule: sugar phosphate isomerase/epimerase;PDBTitle: crystal structure of a sugar phosphate isomerase/epimerase (bdi_1903)2 from parabacteroides distasonis atcc 8503 at 1.85 a resolution
37 c5ikjB_
not modelled
8.7
71
PDB header: transcriptionChain: B: PDB Molecule: cryptic loci regulator protein 1;PDBTitle: structure of clr2 bound to the clr1 c-terminus
38 c2i2oB_
not modelled
8.7
57
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: eif4g-like protein;PDBTitle: crystal structure of an eif4g-like protein from danio rerio
39 c2hk1D_
not modelled
8.6
11
PDB header: isomeraseChain: D: PDB Molecule: d-psicose 3-epimerase;PDBTitle: crystal structure of d-psicose 3-epimerase (dpease) in the presence of2 d-fructose
40 d1ex4a1
not modelled
8.5
71
Fold: SH3-like barrelSuperfamily: DNA-binding domain of retroviral integraseFamily: DNA-binding domain of retroviral integrase
41 d1kpta_
not modelled
8.3
37
Fold: Yeast killer toxinsSuperfamily: Yeast killer toxinsFamily: Virally encoded KP4 toxin
42 d1ihwa_
not modelled
8.3
71
Fold: SH3-like barrelSuperfamily: DNA-binding domain of retroviral integraseFamily: DNA-binding domain of retroviral integrase
43 c6f90A_
not modelled
8.1
20
PDB header: hydrolaseChain: A: PDB Molecule: alpha-1,2-mannosidase, putative;PDBTitle: structure of the family gh92 alpha-mannosidase bt3130 from bacteroides2 thetaiotaomicron in complex with mannoimidazole (mani)
44 c2rbaB_
not modelled
8.0
24
PDB header: hydrolase/dnaChain: B: PDB Molecule: g/t mismatch-specific thymine dna glycosylase;PDBTitle: structure of human thymine dna glycosylase bound to abasic and2 undamaged dna
45 c1wymA_
not modelled
7.6
20
PDB header: structural proteinChain: A: PDB Molecule: transgelin-2;PDBTitle: solution structure of the ch domain of human transgelin-2
46 c4l3uA_
not modelled
7.4
14
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a duf3571 family protein (abaye3784) from2 acinetobacter baumannii aye at 1.95 a resolution
47 c3nzkB_
not modelled
7.3
39
PDB header: hydrolaseChain: B: PDB Molecule: udp-3-o-[3-hydroxymyristoyl] n-acetylglucosaminePDBTitle: structure of lpxc from yersinia enterocolitica complexed with chir0902 inhibitor
48 c5lc5D_
not modelled
7.2
45
PDB header: oxidoreductaseChain: D: PDB Molecule: nadh dehydrogenase [ubiquinone] iron-sulfur protein 2,PDBTitle: structure of mammalian respiratory complex i, class2
49 c3thdD_
not modelled
7.2
35
PDB header: hydrolaseChain: D: PDB Molecule: beta-galactosidase;PDBTitle: crystal structure of human beta-galactosidase in complex with 1-2 deoxygalactonojirimycin
50 d1mpga2
not modelled
7.2
22
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: DNA repair glycosylase, N-terminal domain
51 d1zu2a1
not modelled
7.1
9
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)
52 d1v29b_
not modelled
7.1
33
Fold: SH3-like barrelSuperfamily: Electron transport accessory proteinsFamily: Nitrile hydratase beta chain
53 c3hluA_
not modelled
7.1
31
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein duf2179;PDBTitle: crystal structure of uncharacterized protein conserved in bacteria2 duf2179 from eubacterium ventriosum
54 c3j3vE_
not modelled
7.1
31
PDB header: ribosomeChain: E: PDB Molecule: 50s ribosomal protein l4;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 i-a)
55 c3c66C_
not modelled
7.0
63
PDB header: transferaseChain: C: PDB Molecule: pre-mrna polyadenylation factor fip1;PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
56 c6gcsC_
not modelled
6.9
45
PDB header: oxidoreductaseChain: C: PDB Molecule: 49-kda protein (nucm);PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
57 c3c66D_
not modelled
6.9
63
PDB header: transferaseChain: D: PDB Molecule: pre-mrna polyadenylation factor fip1;PDBTitle: yeast poly(a) polymerase in complex with fip1 residues 80-105
58 c4yl8B_
not modelled
6.9
32
PDB header: protein bindingChain: B: PDB Molecule: protein crumbs;PDBTitle: crystal structure of the crumbs/moesin complex
59 c3j3bq_
not modelled
6.7
6
PDB header: ribosomeChain: Q: PDB Molecule: 60s ribosomal protein l18;PDBTitle: structure of the human 60s ribosomal proteins
60 c6bs7A_
not modelled
6.7
53
PDB header: ligaseChain: A: PDB Molecule: adenylosuccinate synthetase;PDBTitle: crystal structure of adenylosuccinate synthetase from legionella2 pneumophila philadelphia 1
61 d1hh5a_
not modelled
6.5
28
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like
62 c4e8cA_
not modelled
6.4
40
PDB header: hydrolaseChain: A: PDB Molecule: glycosyl hydrolase, family 35;PDBTitle: crystal structure of streptococcal beta-galactosidase in complex with2 galactose
63 d1pc2a_
not modelled
6.3
16
Fold: alpha-alpha superhelixSuperfamily: TPR-likeFamily: Tetratricopeptide repeat (TPR)
64 d2btva_
not modelled
6.3
38
Fold: Reovirus inner layer core protein p3Superfamily: Reovirus inner layer core protein p3Family: Orbivirus core
65 d2cy3a_
not modelled
6.2
30
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like
66 c2btvB_
not modelled
6.2
38
PDB header: virusChain: B: PDB Molecule: protein (vp3 core protein);PDBTitle: atomic model for bluetongue virus (btv) core
67 c3iz5s_
not modelled
6.1
12
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein l18a (l18ae);PDBTitle: localization of the large subunit ribosomal proteins into a 5.5 a2 cryo-em map of triticum aestivum translating 80s ribosome
68 d1hx7a_
not modelled
6.1
32
Fold: Resolvase-likeSuperfamily: Resolvase-likeFamily: gamma,delta resolvase, catalytic domain
69 d1te4a_
not modelled
6.0
20
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: PBS lyase HEAT-like repeat
70 c5mlc2_
not modelled
6.0
36
PDB header: ribosomeChain: 2: PDB Molecule: 50s ribosomal protein l32, chloroplastic;PDBTitle: cryo-em structure of the spinach chloroplast ribosome reveals the2 location of plastid-specific ribosomal proteins and extensions
71 c5f5nA_
not modelled
5.8
50
PDB header: oxidoreductaseChain: A: PDB Molecule: monooxygenase;PDBTitle: the structure of monooxygenase ksta11 in complex with nad and its2 substrate
72 c4ijyA_
not modelled
5.7
55
PDB header: unknown functionChain: A: PDB Molecule: cofj;PDBTitle: crystal structure of the etec secreted protein cofj
73 c4cimQ_
not modelled
5.6
46
PDB header: apoptosisChain: Q: PDB Molecule: bcl-2-like protein 2;PDBTitle: complex of a bcl-w bh3 mutant with a bh3 domain
74 c4cimP_
not modelled
5.6
46
PDB header: apoptosisChain: P: PDB Molecule: bcl-2-like protein 2;PDBTitle: complex of a bcl-w bh3 mutant with a bh3 domain
75 c3h4nB_
not modelled
5.5
27
PDB header: electron transportChain: B: PDB Molecule: cytochrome c7;PDBTitle: ppcd, a cytochrome c7 from geobacter sulfurreducens
76 c2lsqA_
not modelled
5.4
63
PDB header: immune systemChain: A: PDB Molecule: analog of the fragment 197-221 of beta-1 adrenoreceptor;PDBTitle: analog of the fragment 197-221 of beta-1 adrenoreceptor
77 c1lv4A_
not modelled
5.4
50
PDB header: signaling proteinChain: A: PDB Molecule: catestatin;PDBTitle: human catestatin 21-mer
78 c5n4aA_
not modelled
5.4
15
PDB header: transport proteinChain: A: PDB Molecule: intraflagellar transport protein 80;PDBTitle: crystal structure of chlamydomonas ift80
79 c2c2pA_
not modelled
5.3
28
PDB header: hydrolaseChain: A: PDB Molecule: g/u mismatch-specific dna glycosylase;PDBTitle: the crystal structure of mismatch specific uracil-dna2 glycosylase (mug) from deinococcus radiodurans
80 c5ncrA_
not modelled
5.3
22
PDB header: hydrolaseChain: A: PDB Molecule: tyrosine phosphatase;PDBTitle: oh1 from the orf virus: a tyrosine phosphatase that displays distinct2 structural features and triple substrate specificity
81 c3izcs_
not modelled
5.3
6
PDB header: ribosomeChain: S: PDB Molecule: 60s ribosomal protein rpl20 (l18ae);PDBTitle: localization of the large subunit ribosomal proteins into a 6.1 a2 cryo-em map of saccharomyces cerevisiae translating 80s ribosome
82 d1hu3a_
not modelled
5.2
30
Fold: alpha-alpha superhelixSuperfamily: ARM repeatFamily: MIF4G domain-like
83 d1rwja_
not modelled
5.1
40
Fold: Multiheme cytochromesSuperfamily: Multiheme cytochromesFamily: Cytochrome c3-like
84 c6f92B_
not modelled
5.1
23
PDB header: hydrolaseChain: B: PDB Molecule: putative alpha-1,2-mannosidase;PDBTitle: structure of the family gh92 alpha-mannosidase bt3965 from bacteroides2 thetaiotaomicron in complex with mannoimidazole (mani)