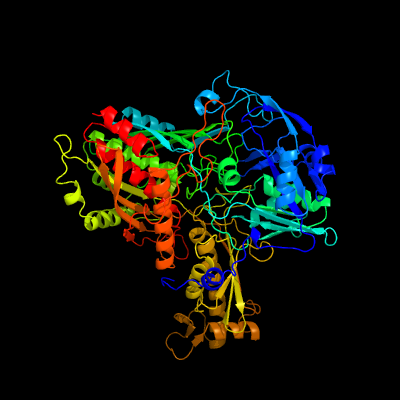
| 1 | c1ffvB_
|
|
 |
100.0 |
59 |
PDB header:hydrolase
Chain: B: PDB Molecule:cutl, molybdoprotein of carbon monoxide
PDBTitle: carbon monoxide dehydrogenase from hydrogenophaga2 pseudoflava
|
|
|
|
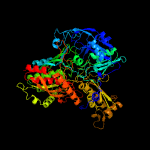
| 2 | c1n62E_
|
|
 |
100.0 |
57 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:carbon monoxide dehydrogenase large chain;
PDBTitle: crystal structure of the mo,cu-co dehydrogenase (codh), n-2 butylisocyanide-bound state
|
|
|
|
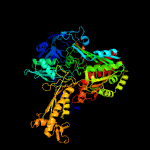
| 3 | c1t3qB_
|
|
 |
100.0 |
38 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:quinoline 2-oxidoreductase large subunit;
PDBTitle: crystal structure of quinoline 2-oxidoreductase from pseudomonas2 putida 86
|
|
|
|
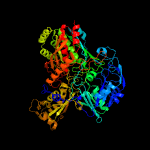
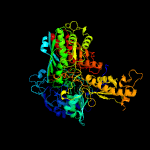
| 4 | c1sb3D_
|
|
 |
100.0 |
28 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:4-hydroxybenzoyl-coa reductase alpha subunit;
PDBTitle: structure of 4-hydroxybenzoyl-coa reductase from thauera2 aromatica
|
|
|
|
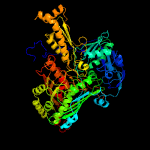
| 5 | c1wygA_
|
|
 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of a rat xanthine dehydrogenase triple mutant2 (c535a, c992r and c1324s)
|
|
|
|
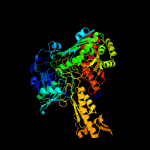
| 6 | c2w54F_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:xanthine dehydrogenase;
PDBTitle: crystal structure of xanthine dehydrogenase from rhodobacter2 capsulatus in complex with bound inhibitor pterin-6-aldehyde
|
|
|
|
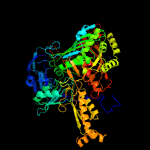
| 7 | c4uhxA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde oxidase;
PDBTitle: human aldehyde oxidase in complex with phthalazine and thioridazine
|
|
|
|
| 8 | c5y6qC_
|
|
 |
100.0 |
26 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:aldehyde oxidase large subunit;
PDBTitle: crystal structure of an aldehyde oxidase from methylobacillus sp.2 ky4400
|
|
|
|
| 9 | c3zyvA_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aox3;
PDBTitle: crystal structure of the mouse liver aldehyde oxidase 3 (maox3)
|
|
|
|
| 10 | c3eubL_
|
|
 |
100.0 |
21 |
PDB header:oxidoreductase
Chain: L: PDB Molecule:xanthine dehydrogenase/oxidase;
PDBTitle: crystal structure of desulfo-xanthine oxidase with xanthine
|
|
|
|
| 11 | c5g5hC_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:putative xanthine dehydrogenase yagr molybdenum-binding
PDBTitle: escherichia coli periplasmic aldehyde oxidase r440h mutant
|
|
|
|
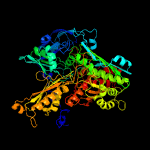
| 12 | c1vlbA_
|
|
 |
100.0 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde oxidoreductase;
PDBTitle: structure refinement of the aldehyde oxidoreductase from2 desulfovibrio gigas at 1.28 a
|
|
|
|
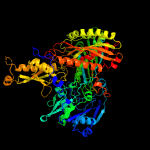
| 13 | c1dgjA_
|
|
 |
100.0 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:aldehyde oxidoreductase;
PDBTitle: crystal structure of the aldehyde oxidoreductase from2 desulfovibrio desulfuricans atcc 27774
|
|
|
|
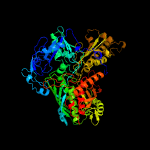
| 14 | c4zohA_
|
|
 |
100.0 |
36 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:putative oxidoreductase molybdopterin-binding subunit;
PDBTitle: crystal structure of glyceraldehyde oxidoreductase
|
|
|
|
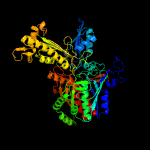
| 15 | d1n62b2
|
|
 |
100.0 |
57 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
|
|
|
| 16 | d1ffvb2
|
|
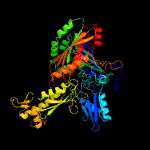 |
100.0 |
61 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
|
|
|
| 17 | d1rm6a2
|
|
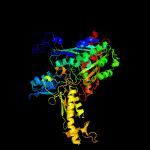 |
100.0 |
26 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
|
|
|
| 18 | d1t3qb2
|
|
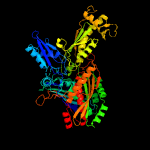 |
100.0 |
37 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
|
|
|
| 19 | d1jrob2
|
|
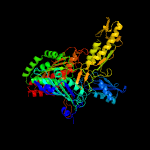 |
100.0 |
22 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
|
|
|
| 20 | d1v97a5
|
|
 |
100.0 |
20 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
|
|
|
| 21 | d1vlba4 |
|
not modelled |
100.0 |
25 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
|
|
| 22 | d1dgja4 |
|
not modelled |
100.0 |
24 |
Fold:Molybdenum cofactor-binding domain
Superfamily:Molybdenum cofactor-binding domain
Family:Molybdenum cofactor-binding domain |
|
|
| 23 | c3hrdE_ |
|
not modelled |
100.0 |
26 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:nicotinate dehydrogenase large molybdopterin
PDBTitle: crystal structure of nicotinate dehydrogenase
|
|
|
| 24 | c3hrdF_ |
|
not modelled |
100.0 |
31 |
PDB header:oxidoreductase
Chain: F: PDB Molecule:nicotinate dehydrogenase medium molybdopterin
PDBTitle: crystal structure of nicotinate dehydrogenase
|
|
|
| 25 | d1t3qb1 |
|
not modelled |
100.0 |
43 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 26 | d1ffvb1 |
|
not modelled |
100.0 |
54 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 27 | d1n62b1 |
|
not modelled |
100.0 |
58 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 28 | d1v97a3 |
|
not modelled |
100.0 |
24 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 29 | d3b9jc1 |
|
not modelled |
100.0 |
26 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 30 | d1rm6a1 |
|
not modelled |
100.0 |
37 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 31 | d1jrob1 |
|
not modelled |
100.0 |
33 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 32 | d1vlba3 |
|
not modelled |
100.0 |
33 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 33 | d1dgja3 |
|
not modelled |
100.0 |
31 |
Fold:alpha/beta-Hammerhead
Superfamily:CO dehydrogenase molybdoprotein N-domain-like
Family:CO dehydrogenase molybdoprotein N-domain-like |
|
|
| 34 | d1gd0a_ |
|
not modelled |
75.3 |
15 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
| 35 | d2gdga1 |
|
not modelled |
74.4 |
14 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
| 36 | d1hfoa_ |
|
not modelled |
74.2 |
13 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
| 37 | d1uiza_ |
|
not modelled |
74.0 |
11 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
| 38 | c5un4C_ |
|
not modelled |
72.8 |
12 |
PDB header:hydrolase
Chain: C: PDB Molecule:4-oxalocrotonate tautomerase;
PDBTitle: crystal structure of native fused 4-ot
|
|
|
| 39 | c4dh4A_ |
|
not modelled |
70.6 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:mif;
PDBTitle: macrophage migration inhibitory factor toxoplasma gondii
|
|
|
| 40 | d1fima_ |
|
not modelled |
69.4 |
14 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
| 41 | d1yh5a1 |
|
not modelled |
69.0 |
4 |
Fold:YggU-like
Superfamily:YggU-like
Family:YggU-like |
|
|
| 42 | c6cuqB_ |
|
not modelled |
67.2 |
11 |
PDB header:cytokine
Chain: B: PDB Molecule:macrophage migration inhibitory factor-like protein;
PDBTitle: crystal structure of macrophage migration inhibitory factor-like2 protein (ehmif) from entamoeba histolytica
|
|
|
| 43 | d1dgja1 |
|
not modelled |
65.7 |
8 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
|
|
| 44 | c2xczA_ |
|
not modelled |
65.6 |
10 |
PDB header:immune system
Chain: A: PDB Molecule:possible atls1-like light-inducible protein;
PDBTitle: crystal structure of macrophage migration inhibitory factor homologue2 from prochlorococcus marinus
|
|
|
| 45 | c2os5C_ |
|
not modelled |
64.7 |
10 |
PDB header:cytokine
Chain: C: PDB Molecule:acemif;
PDBTitle: macrophage migration inhibitory factor from ancylostoma ceylanicum
|
|
|
| 46 | c3gacD_ |
|
not modelled |
64.2 |
19 |
PDB header:cytokine
Chain: D: PDB Molecule:macrophage migration inhibitory factor-like
PDBTitle: structure of mif with hpp
|
|
|
| 47 | c3b64A_ |
|
not modelled |
62.4 |
13 |
PDB header:cytokine
Chain: A: PDB Molecule:macrophage migration inhibitory factor-like
PDBTitle: macrophage migration inhibitory factor (mif) from2 /leishmania major
|
|
|
| 48 | c2rghA_ |
|
not modelled |
61.5 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
|
|
| 49 | d2dt5a1 |
|
not modelled |
60.8 |
13 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Transcriptional repressor Rex, N-terminal domain |
|
|
| 50 | c5unqF_ |
|
not modelled |
60.8 |
5 |
PDB header:hydrolase
Chain: F: PDB Molecule:putative tautomerase;
PDBTitle: crystal structure of pt0534 inactivated by 2-oxo-3-pentynoate
|
|
|
| 51 | d1dpta_ |
|
not modelled |
59.0 |
12 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MIF-related |
|
|
| 52 | c4lkbA_ |
|
not modelled |
58.3 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:hypothetical protein alr4568/putative 4-oxalocrotonate
PDBTitle: crystal structure of a putative 4-oxalocrotonate tautomerase from2 nostoc sp. pcc 7120
|
|
|
| 53 | c3fwtA_ |
|
not modelled |
57.0 |
14 |
PDB header:cytokine
Chain: A: PDB Molecule:macrophage migration inhibitory factor-like
PDBTitle: crystal structure of leishmania major mif2
|
|
|
| 54 | d1otfa_ |
|
not modelled |
52.6 |
14 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:4-oxalocrotonate tautomerase-like |
|
|
| 55 | c3abfB_ |
|
not modelled |
52.3 |
11 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate tautomerase;
PDBTitle: crystal structure of a 4-oxalocrotonate tautomerase homologue2 (tthb242)
|
|
|
| 56 | c3ry0A_ |
|
not modelled |
51.6 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:putative tautomerase;
PDBTitle: crystal structure of tomn, a 4-oxalocrotonate tautomerase homologue in2 tomaymycin biosynthetic pathway
|
|
|
| 57 | d1mwwa_ |
|
not modelled |
51.1 |
16 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:Hypothetical protein HI1388.1 |
|
|
| 58 | c2op8A_ |
|
not modelled |
51.0 |
14 |
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase ywhb;
PDBTitle: crystal structure of ywhb- homologue of 4-oxalocrotonate tautomerase
|
|
|
| 59 | c4fazB_ |
|
not modelled |
49.2 |
19 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate isomerase protein;
PDBTitle: kinetic and structural characterization of the 4-oxalocrotonate2 tautomerase isozymes from methylibium petroleiphilum
|
|
|
| 60 | c4fdxB_ |
|
not modelled |
49.0 |
14 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonase tautomerase isozyme;
PDBTitle: kinetic and structural characterization of the 4-oxalocrotonate2 tautomerase isozymes from methylibium petroleiphilum
|
|
|
| 61 | d1vlba1 |
|
not modelled |
48.9 |
11 |
Fold:CO dehydrogenase ISP C-domain like
Superfamily:CO dehydrogenase ISP C-domain like
Family:CO dehydrogenase ISP C-domain like |
|
|
| 62 | d1bjpa_ |
|
not modelled |
48.4 |
14 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:4-oxalocrotonate tautomerase-like |
|
|
| 63 | d2aala1 |
|
not modelled |
47.3 |
18 |
Fold:Tautomerase/MIF
Superfamily:Tautomerase/MIF
Family:MSAD-like |
|
|
| 64 | c2dt5A_ |
|
not modelled |
46.8 |
12 |
PDB header:dna binding protein
Chain: A: PDB Molecule:at-rich dna-binding protein;
PDBTitle: crystal structure of ttha1657 (at-rich dna-binding protein) from2 thermus thermophilus hb8
|
|
|
| 65 | c2lfcA_ |
|
not modelled |
46.1 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:fumarate reductase, flavoprotein subunit;
PDBTitle: solution nmr structure of fumarate reductase flavoprotein subunit from2 lactobacillus plantarum, northeast structural genomics consortium3 target lpr145j
|
|
|
| 66 | c3n2oA_ |
|
not modelled |
44.1 |
22 |
PDB header:lyase
Chain: A: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: x-ray crystal structure of arginine decarboxylase complexed with2 arginine from vibrio vulnificus
|
|
|
| 67 | c3m20A_ |
|
not modelled |
43.7 |
17 |
PDB header:isomerase
Chain: A: PDB Molecule:4-oxalocrotonate tautomerase, putative;
PDBTitle: crystal structure of dmpi from archaeoglobus fulgidus determined to2 2.37 angstroms resolution
|
|
|
| 68 | c2kwpA_ |
|
not modelled |
43.6 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:transcription elongation protein nusa;
PDBTitle: solution structure of the aminoterminal domain of e. coli nusa
|
|
|
| 69 | d1knwa2 |
|
not modelled |
43.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:PLP-binding barrel
Family:Alanine racemase-like, N-terminal domain |
|
|
| 70 | c3mlcC_ |
|
not modelled |
42.1 |
12 |
PDB header:isomerase
Chain: C: PDB Molecule:fg41 malonate semialdehyde decarboxylase;
PDBTitle: crystal structure of fg41msad inactivated by 3-chloropropiolate
|
|
|
| 71 | d1hh2p4 |
|
not modelled |
42.0 |
36 |
Fold:Transcription factor NusA, N-terminal domain
Superfamily:Transcription factor NusA, N-terminal domain
Family:Transcription factor NusA, N-terminal domain |
|
|
| 72 | c4aibC_ |
|
not modelled |
40.9 |
13 |
PDB header:lyase
Chain: C: PDB Molecule:ornithine decarboxylase;
PDBTitle: crystal structure of ornithine decarboxylase from entamoeba2 histolytica.
|
|
|
| 73 | c3mb2G_ |
|
not modelled |
39.1 |
9 |
PDB header:isomerase
Chain: G: PDB Molecule:4-oxalocrotonate tautomerase family enzyme - alpha subunit;
PDBTitle: kinetic and structural characterization of a heterohexamer 4-2 oxalocrotonate tautomerase from chloroflexus aurantiacus j-10-fl:3 implications for functional and structural diversity in the4 tautomerase superfamily
|
|
|
| 74 | c2mt4A_ |
|
not modelled |
37.5 |
27 |
PDB header:transcription
Chain: A: PDB Molecule:transcription termination/antitermination protein nusa;
PDBTitle: solution structure of the n-terminal domain of nusa from b. subtilis
|
|
|
| 75 | c5wc3S_ |
|
not modelled |
37.2 |
20 |
PDB header:protein transport
Chain: S: PDB Molecule:spoiiiag, stage iii sporulation engulfment assemblyprotein;
PDBTitle: spoiiiag
|
|
|
| 76 | c2rgoA_ |
|
not modelled |
37.1 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alpha-glycerophosphate oxidase;
PDBTitle: structure of alpha-glycerophosphate oxidase from2 streptococcus sp.: a template for the mitochondrial alpha-3 glycerophosphate dehydrogenase
|
|
|
| 77 | c2hlhA_ |
|
not modelled |
36.6 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:nodulation fucosyltransferase;
PDBTitle: crystal structure of fucosyltransferase nodz from bradyrhizobium
|
|
|
| 78 | c2ormA_ |
|
not modelled |
36.6 |
20 |
PDB header:isomerase
Chain: A: PDB Molecule:probable tautomerase hp0924;
PDBTitle: crystal structure of the 4-oxalocrotonate tautomerase homologue dmpi2 from helicobacter pylori.
|
|
|
| 79 | c2x4kB_ |
|
not modelled |
36.3 |
16 |
PDB header:isomerase
Chain: B: PDB Molecule:4-oxalocrotonate tautomerase;
PDBTitle: crystal structure of sar1376, a putative 4-oxalocrotonate2 tautomerase from the methicillin-resistant staphylococcus3 aureus (mrsa)
|
|
|
| 80 | c4gisB_ |
|
not modelled |
36.2 |
20 |
PDB header:lyase
Chain: B: PDB Molecule:enolase;
PDBTitle: crystal structure of an enolase family member from vibrio harveyi2 (efi-target 501692) with homology to mannonate dehydratase, with mg,3 glycerol and dicarboxylates bound (mixed loops, space group i4122)
|
|
|
| 81 | c3t5sA_ |
|
not modelled |
35.4 |
14 |
PDB header:immune system
Chain: A: PDB Molecule:macrophage migration inhibitory factor;
PDBTitle: structure of macrophage migration inhibitory factor from giardia2 lamblia
|
|
|
| 82 | d1d4ca3 |
|
not modelled |
35.4 |
29 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
|
|
| 83 | d1o6za2 |
|
not modelled |
34.3 |
17 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
|
|
| 84 | d1y0pa3 |
|
not modelled |
33.5 |
18 |
Fold:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Superfamily:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain
Family:Succinate dehydrogenase/fumarate reductase flavoprotein, catalytic domain |
|
|
| 85 | c3da1A_ |
|
not modelled |
33.2 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol-3-phosphate dehydrogenase;
PDBTitle: x-ray structure of the glycerol-3-phosphate dehydrogenase2 from bacillus halodurans complexed with fad. northeast3 structural genomics consortium target bhr167.
|
|
|
| 86 | c5uifC_ |
|
not modelled |
33.1 |
10 |
PDB header:hydrolase
Chain: C: PDB Molecule:ps01740;
PDBTitle: crystal structure of native ps01740
|
|
|
| 87 | c3ketA_ |
|
not modelled |
32.2 |
24 |
PDB header:transcription/dna
Chain: A: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: crystal structure of a rex-family transcriptional regulatory protein2 from streptococcus agalactiae bound to a palindromic operator
|
|
|
| 88 | c3nzqB_ |
|
not modelled |
30.1 |
27 |
PDB header:lyase
Chain: B: PDB Molecule:biosynthetic arginine decarboxylase;
PDBTitle: crystal structure of biosynthetic arginine decarboxylase adc (spea)2 from escherichia coli, northeast structural genomics consortium3 target er600
|
|
|
| 89 | d1nera_ |
|
not modelled |
29.8 |
32 |
Fold:lambda repressor-like DNA-binding domains
Superfamily:lambda repressor-like DNA-binding domains
Family:Phage repressors |
|
|
| 90 | c3nzpA_ |
|
not modelled |
29.4 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:arginine decarboxylase;
PDBTitle: crystal structure of the biosynthetic arginine decarboxylase spea from2 campylobacter jejuni, northeast structural genomics consortium target3 br53
|
|
|
| 91 | c2lteA_ |
|
not modelled |
26.4 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:specialized acyl carrier protein;
PDBTitle: solution nmr structure of the specialized acyl carrier protein pa33342 (apo) from pseudomonas aeruginosa, northeast structural genomics3 consortium target pat415
|
|
|
| 92 | d1t0aa_ |
|
not modelled |
24.0 |
18 |
Fold:Bacillus chorismate mutase-like
Superfamily:IpsF-like
Family:IpsF-like |
|
|
| 93 | c6aeoA_ |
|
not modelled |
23.9 |
24 |
PDB header:protein transport
Chain: A: PDB Molecule:maltose/maltodextrin-binding periplasmic protein,tssl;
PDBTitle: tssl periplasmic domain
|
|
|
| 94 | c1l0oC_ |
|
not modelled |
23.4 |
29 |
PDB header:protein binding
Chain: C: PDB Molecule:sigma factor;
PDBTitle: crystal structure of the bacillus stearothermophilus anti-2 sigma factor spoiiab with the sporulation sigma factor3 sigmaf
|
|
|
| 95 | d1l0oc_ |
|
not modelled |
23.4 |
29 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:Sigma3 and sigma4 domains of RNA polymerase sigma factors
Family:Sigma4 domain |
|
|
| 96 | c1w36E_ |
|
not modelled |
23.2 |
22 |
PDB header:recombination
Chain: E: PDB Molecule:exodeoxyribonuclease v beta chain;
PDBTitle: recbcd:dna complex
|
|
|
| 97 | c1vraA_ |
|
not modelled |
22.6 |
16 |
PDB header:transferase
Chain: A: PDB Molecule:arginine biosynthesis bifunctional protein argj;
PDBTitle: crystal structure of arginine biosynthesis bifunctional protein argj2 (10175521) from bacillus halodurans at 2.00 a resolution
|
|
|
| 98 | d2fmra_ |
|
not modelled |
22.4 |
18 |
Fold:Eukaryotic type KH-domain (KH-domain type I)
Superfamily:Eukaryotic type KH-domain (KH-domain type I)
Family:Eukaryotic type KH-domain (KH-domain type I) |
|
|
| 99 | d2f2ab2 |
|
not modelled |
22.3 |
17 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:GatB/GatE catalytic domain-like |
|
|
| 100 | c3wg9D_ |
|
not modelled |
22.2 |
13 |
PDB header:transcription
Chain: D: PDB Molecule:redox-sensing transcriptional repressor rex;
PDBTitle: crystal structure of rsp, a rex-family repressor
|
|
|
| 101 | d7mdha2 |
|
not modelled |
21.7 |
11 |
Fold:LDH C-terminal domain-like
Superfamily:LDH C-terminal domain-like
Family:Lactate & malate dehydrogenases, C-terminal domain |
|
|
| 102 | c4w8jA_ |
|
not modelled |
21.3 |
15 |
PDB header:toxin
Chain: A: PDB Molecule:pesticidal crystal protein cry1ac;
PDBTitle: structure of the full-length insecticidal protein cry1ac reveals2 intriguing details of toxin packaging into in vivo formed crystals
|
|
|
| 103 | c5ijwA_ |
|
not modelled |
20.8 |
24 |
PDB header:isomerase
Chain: A: PDB Molecule:glutamate racemase;
PDBTitle: glutamate racemase (muri) from mycobacterium smegmatis with bound d-2 glutamate, 1.8 angstrom resolution, x-ray diffraction
|
|
|
| 104 | c1af7A_ |
|
not modelled |
20.8 |
24 |
PDB header:methyltransferase
Chain: A: PDB Molecule:chemotaxis receptor methyltransferase cher;
PDBTitle: cher from salmonella typhimurium
|
|
|