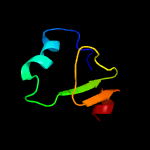
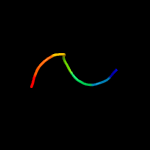
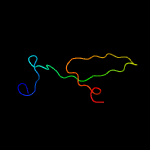
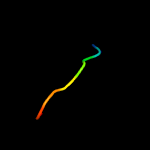
1 d3c0na2
46.5
25
Fold: Aerolisin/ETX pore-forming domainSuperfamily: Aerolisin/ETX pore-forming domainFamily: (Pro)aerolysin, pore-forming lobe
2 c1urzC_
32.5
18
PDB header: virus/viral proteinChain: C: PDB Molecule: envelope protein;PDBTitle: low ph induced, membrane fusion conformation of the2 envelope protein of tick-borne encephalitis virus
3 c4iaoB_
24.7
37
PDB header: hydrolase/transcriptionChain: B: PDB Molecule: nad-dependent histone deacetylase sir2;PDBTitle: crystal structure of sir2 c543s mutant in complex with sid domain of2 sir4
4 d1ok8a2
14.8
20
Fold: Viral glycoprotein, central and dimerisation domainsSuperfamily: Viral glycoprotein, central and dimerisation domainsFamily: Viral glycoprotein, central and dimerisation domains
5 d1qcsa1
13.3
27
Fold: Double psi beta-barrelSuperfamily: ADC-likeFamily: Cdc48 N-terminal domain-like
6 d1svba2
11.7
21
Fold: Viral glycoprotein, central and dimerisation domainsSuperfamily: Viral glycoprotein, central and dimerisation domainsFamily: Viral glycoprotein, central and dimerisation domains
7 c5ireA_
11.5
22
PDB header: virusChain: A: PDB Molecule: e protein;PDBTitle: the cryo-em structure of zika virus
8 c6bi6A_
8.4
24
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein yejg;PDBTitle: solution nmr structure of uncharacterized protein yejg
9 d1i40a_
7.3
29
Fold: OB-foldSuperfamily: Inorganic pyrophosphataseFamily: Inorganic pyrophosphatase
10 c4o4dA_
7.0
23
PDB header: transferaseChain: A: PDB Molecule: inositol hexakisphosphate kinase;PDBTitle: crystal structure of an inositol hexakisphosphate kinase ehip6ka in2 complexed with atp and ins(1,4,5)p3
11 c2hjhB_
7.0
32
PDB header: hydrolaseChain: B: PDB Molecule: nad-dependent histone deacetylase sir2;PDBTitle: crystal structure of the sir2 deacetylase
12 d2erla_
7.0
86
Fold: Protozoan pheromone-likeSuperfamily: Protozoan pheromone proteinsFamily: Protozoan pheromone proteins
13 d2cu9a1
6.8
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ASF1-likeFamily: ASF1-like
14 d1roca_
6.6
23
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ASF1-likeFamily: ASF1-like
15 c4drbC_
6.5
20
PDB header: dna binding protein/protein bindingChain: C: PDB Molecule: fanconi anemia group m protein;PDBTitle: the crystal structure of fancm bound mhf complex
16 c3uajA_
6.5
21
PDB header: viral protein/immune systemChain: A: PDB Molecule: envelope protein;PDBTitle: crystal structure of the envelope glycoprotein ectodomain from dengue2 virus serotype 4 in complex with the fab fragment of the chimpanzee3 monoclonal antibody 5h2
17 d2z1ca1
6.2
33
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like
18 c2idcA_
6.1
23
PDB header: replication/chaperoneChain: A: PDB Molecule: anti-silencing protein 1 and histone h3 chimera;PDBTitle: structure of the histone h3-asf1 chaperone interaction
19 c3j21R_
5.9
36
PDB header: ribosomeChain: R: PDB Molecule: 50s ribosomal protein l21e;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
20 c5vqjA_
5.9
28
PDB header: hydrolaseChain: A: PDB Molecule: exo-beta-1,4-xylanase;PDBTitle: discovery of a first gh11 exo-1,4-beta-xylanase from a diverse2 microbial sugar cane bagasse composting community
21 c3lmaC_
not modelled
5.9
37
PDB header: membrane proteinChain: C: PDB Molecule: stage v sporulation protein ad (spovad);PDBTitle: crystal structure of the stage v sporulation protein ad (spovad) from2 bacillus licheniformis. northeast structural genomics consortium3 target bir6.
22 d2ot2a1
not modelled
5.7
44
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like
23 c2i32A_
not modelled
5.7
27
PDB header: replication chaperoneChain: A: PDB Molecule: anti-silencing factor 1 paralog a;PDBTitle: structure of a human asf1a-hira complex and insights into specificity2 of histone chaperone complex assembly
24 d2i32a1
not modelled
5.7
27
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ASF1-likeFamily: ASF1-like
25 d1vmba_
not modelled
5.6
25
Fold: Ferredoxin-likeSuperfamily: Ribosomal protein S6Family: Ribosomal protein S6
26 c1vmbA_
not modelled
5.6
25
PDB header: translationChain: A: PDB Molecule: 30s ribosomal protein s6;PDBTitle: crystal structure of 30s ribosomal protein s6 (tm0603) from thermotoga2 maritima at 1.70 a resolution
27 d1loua_
not modelled
5.6
13
Fold: Ferredoxin-likeSuperfamily: Ribosomal protein S6Family: Ribosomal protein S6
28 c3ld3A_
not modelled
5.4
33
PDB header: hydrolaseChain: A: PDB Molecule: inorganic pyrophosphatase;PDBTitle: crystal structure of inorganic phosphatase from anaplasma2 phagocytophilum at 1.75a resolution
29 d1pz8a_
not modelled
5.4
38
Fold: Concanavalin A-like lectins/glucanasesSuperfamily: Concanavalin A-like lectins/glucanasesFamily: Laminin G-like module
30 d1j8fa_
not modelled
5.3
22
Fold: DHS-like NAD/FAD-binding domainSuperfamily: DHS-like NAD/FAD-binding domainFamily: Sir2 family of transcriptional regulators
31 d1v3ea_
not modelled
5.2
18
Fold: 6-bladed beta-propellerSuperfamily: SialidasesFamily: Sialidases (neuraminidases)
32 c4bg7A_
not modelled
5.2
39
PDB header: replicationChain: A: PDB Molecule: putative transcriptional coactivator p15;PDBTitle: bacteriophage t5 homolog of the eukaryotic transcription coactivator2 pc4 implicated in recombination-dependent dna replication
33 d3d3ra1
not modelled
5.1
67
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like