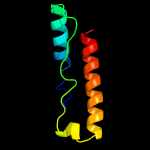
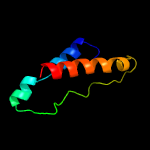
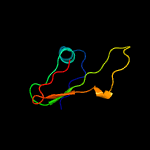
1 c2miiA_
93.4
18
PDB header: protein bindingChain: A: PDB Molecule: penicillin-binding protein activator lpob;PDBTitle: nmr structure of e. coli lpob
2 d1muwa_
47.5
15
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
3 c5buvC_
46.5
14
PDB header: isomeraseChain: C: PDB Molecule: putative epimerase;PDBTitle: x-ray structure of wbca from yersinia enterocolitica
4 c6c46E_
45.2
21
PDB header: isomeraseChain: E: PDB Molecule: dtdp-4-dehydrorhamnose 3,5-epimerase;PDBTitle: crystal structure of dtdp-4-dehydrorhamnose 3,5-epimerase from2 elizabethkingia anophelis nuhp1
5 d1xlma_
44.5
18
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
6 d1dzra_
43.9
16
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
7 d1m0da_
43.9
20
Fold: Restriction endonuclease-likeSuperfamily: Restriction endonuclease-likeFamily: Endonuclease I (Holliday junction resolvase)
8 d1e7la1
43.2
19
Fold: LEM/SAP HeH motifSuperfamily: Recombination endonuclease VII, C-terminal and dimerization domainsFamily: Recombination endonuclease VII, C-terminal and dimerization domains
9 d1wlta1
41.1
19
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
10 c3rykB_
39.2
23
PDB header: isomeraseChain: B: PDB Molecule: dtdp-4-dehydrorhamnose 3,5-epimerase;PDBTitle: 1.63 angstrom resolution crystal structure of dtdp-4-dehydrorhamnose2 3,5-epimerase (rfbc) from bacillus anthracis str. ames with tdp and3 ppi bound
11 d2ixca1
38.3
26
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
12 d2c0za1
37.5
26
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
13 c1upiA_
36.6
26
PDB header: epimeraseChain: A: PDB Molecule: dtdp-4-dehydrorhamnose 3,5-epimerase;PDBTitle: mycobacterium tuberculosis rmlc epimerase (rv3465)
14 c4hmzD_
36.2
28
PDB header: unknown functionChain: D: PDB Molecule: putative 3-epimerase in d-allose pathway;PDBTitle: crystal structure of chmj, a 3'-monoepimerase from streptomyces2 bikiniensis in complex with dtdp-quinovose
15 c2c0zA_
36.1
26
PDB header: isomeraseChain: A: PDB Molecule: novw;PDBTitle: the 1.6 a resolution crystal structure of novw: a 4-keto-6-2 deoxy sugar epimerase from the novobiocin biosynthetic3 gene cluster of streptomyces spheroides
16 d1oi6a_
33.7
25
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
17 c4nl6C_
32.4
18
PDB header: splicingChain: C: PDB Molecule: survival motor neuron protein;PDBTitle: structure of the full-length form of the protein smn found in healthy2 patients
18 c4k3zA_
31.0
6
PDB header: oxidoreductaseChain: A: PDB Molecule: d-erythrulose 4-phosphate dehydrogenase;PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
19 d1bxca_
27.5
10
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
20 c6ndrA_
26.9
18
PDB header: sugar binding proteinChain: A: PDB Molecule: dtdp-4-dehydrorhamnose 3,5-epimerase;PDBTitle: crystal structure of dtdp-6-deoxy-d-glucose-3,5-epimerase rmlc from2 legionella pneumophila philadelphia 1 in complex with dtdp-4-keto-l-3 rhamnose
21 c3g9hA_
not modelled
25.5
15
PDB header: endocytosisChain: A: PDB Molecule: suppressor of yeast profilin deletion;PDBTitle: crystal structure of the c-terminal mu homology domain of2 syp1
22 d1qt1a_
not modelled
24.3
15
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
23 c2kngA_
not modelled
22.5
33
PDB header: dna binding proteinChain: A: PDB Molecule: protein lsr2;PDBTitle: solution structure of c-domain of lsr2
24 d2glka1
not modelled
21.4
15
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
25 c4xtnJ_
not modelled
21.4
12
PDB header: membrane proteinChain: J: PDB Molecule: sodium pumping rhodopsin;PDBTitle: crystal structure of the light-driven sodium pump kr2 in the2 pentameric red form, ph 4.9
26 c2yopB_
not modelled
21.0
16
PDB header: apoptosisChain: B: PDB Molecule: protein fam3b;PDBTitle: long wavelength s-sad structure of fam3b pander
27 c3d3rA_
not modelled
19.2
13
PDB header: chaperoneChain: A: PDB Molecule: hydrogenase assembly chaperone hypc/hupf;PDBTitle: crystal structure of the hydrogenase assembly chaperone hypc/hupf2 family protein from shewanella oneidensis mr-1
28 d2ixha1
not modelled
18.8
21
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
29 c2jagA_
not modelled
18.6
13
PDB header: membrane proteinChain: A: PDB Molecule: halorhodopsin;PDBTitle: l1-intermediate of halorhodopsin t203v
30 d2z1ca1
not modelled
18.0
13
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like
31 d1qzza2
not modelled
17.9
11
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Plant O-methyltransferase, C-terminal domain
32 d1e12a_
not modelled
17.7
13
Fold: Family A G protein-coupled receptor-likeSuperfamily: Family A G protein-coupled receptor-likeFamily: Bacteriorhodopsin-like
33 d1uaza_
not modelled
17.4
10
Fold: Family A G protein-coupled receptor-likeSuperfamily: Family A G protein-coupled receptor-likeFamily: Bacteriorhodopsin-like
34 d3d3ra1
not modelled
17.4
13
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like
35 d1xioa_
not modelled
16.6
9
Fold: Family A G protein-coupled receptor-likeSuperfamily: Family A G protein-coupled receptor-likeFamily: Bacteriorhodopsin-like
36 c1xioA_
not modelled
16.6
9
PDB header: signaling proteinChain: A: PDB Molecule: anabaena sensory rhodopsin;PDBTitle: anabaena sensory rhodopsin
37 c5gzxD_
not modelled
16.4
18
PDB header: hydrolaseChain: D: PDB Molecule: (r)-2-haloacid dehalogenase;PDBTitle: the complex structure of d-2-haloacid dehalogenase mutant with d-2-cpa
38 c5azdA_
not modelled
16.1
24
PDB header: transport proteinChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: crystal structure of thermophilic rhodopsin.
39 c6eyuA_
not modelled
15.2
15
PDB header: membrane proteinChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: crystal structure of the inward h(+) pump xenorhodopsin
40 c3re2A_
not modelled
15.1
19
PDB header: unknown functionChain: A: PDB Molecule: predicted protein;PDBTitle: crystal structure of menin reveals the binding site for mixed lineage2 leukemia (mll) protein
41 d1h2sa_
not modelled
14.7
18
Fold: Family A G protein-coupled receptor-likeSuperfamily: Family A G protein-coupled receptor-likeFamily: Bacteriorhodopsin-like
42 c3a7kD_
not modelled
14.7
15
PDB header: membrane proteinChain: D: PDB Molecule: halorhodopsin;PDBTitle: crystal structure of halorhodopsin from natronomonas2 pharaonis
43 d1xima_
not modelled
14.7
11
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
44 c2x7lP_
not modelled
14.6
33
PDB header: immune systemChain: P: PDB Molecule: protein rev;PDBTitle: implications of the hiv-1 rev dimer structure at 3.2a resolution for2 multimeric binding to the rev response element
45 c2kncB_
not modelled
14.2
11
PDB header: cell adhesionChain: B: PDB Molecule: integrin beta-3;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
46 c3ddlB_
not modelled
13.9
25
PDB header: transport proteinChain: B: PDB Molecule: xanthorhodopsin;PDBTitle: crystallographic structure of xanthorhodopsin, a light-driven ion pump2 with dual chromophore
47 c3u88B_
not modelled
13.6
32
PDB header: transcriptionChain: B: PDB Molecule: menin;PDBTitle: crystal structure of human menin in complex with mll1 and ledgf
48 c3mk7F_
not modelled
13.1
23
PDB header: oxidoreductaseChain: F: PDB Molecule: cytochrome c oxidase, cbb3-type, subunit p;PDBTitle: the structure of cbb3 cytochrome oxidase
49 c4jr8A_
not modelled
12.9
12
PDB header: proton transportChain: A: PDB Molecule: cruxrhodopsin-3;PDBTitle: crystal structure of cruxrhodopsin-3 from haloarcula vallismortis at2 2.3 angstrom resolution
50 c2ghjD_
not modelled
12.7
32
PDB header: structural proteinChain: D: PDB Molecule: 50s ribosomal protein l20;PDBTitle: crystal structure of folded and partially unfolded forms of aquifex2 aeolicus ribosomal protein l20
51 c3ug9A_
not modelled
12.4
8
PDB header: membrane proteinChain: A: PDB Molecule: archaeal-type opsin 1, archaeal-type opsin 2;PDBTitle: crystal structure of the closed state of channelrhodopsin
52 c4fbzA_
not modelled
12.4
9
PDB header: membrane proteinChain: A: PDB Molecule: deltarhodopsin;PDBTitle: crystal structure of deltarhodopsin from haloterrigena thermotolerans
53 d2ot2a1
not modelled
12.0
20
Fold: OB-foldSuperfamily: HupF/HypC-likeFamily: HupF/HypC-like
54 d1ep0a_
not modelled
12.0
20
Fold: Double-stranded beta-helixSuperfamily: RmlC-like cupinsFamily: dTDP-sugar isomerase
55 d1gz0a2
not modelled
11.8
14
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: RNA 2'-O ribose methyltransferase substrate binding domain
56 c2n1pA_
not modelled
11.7
30
PDB header: viral proteinChain: A: PDB Molecule: non-structural protein 5b, ns5b;PDBTitle: structure of the c-terminal membrane domain of hcv ns5b protein
57 c4qidB_
not modelled
11.6
12
PDB header: membrane proteinChain: B: PDB Molecule: bacteriorhodopsin-i;PDBTitle: crystal structure of haloquadratum walsbyi bacteriorhodopsin
58 d1tw3a2
not modelled
11.4
9
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Plant O-methyltransferase, C-terminal domain
59 c3kdpG_
not modelled
11.2
21
PDB header: hydrolaseChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
60 c3kdpH_
not modelled
11.2
21
PDB header: hydrolaseChain: H: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of the sodium-potassium pump
61 c3lphD_
not modelled
11.2
33
PDB header: viral proteinChain: D: PDB Molecule: protein rev;PDBTitle: crystal structure of the hiv-1 rev dimer
62 d1z01a1
not modelled
11.1
16
Fold: ISP domainSuperfamily: ISP domainFamily: Ring hydroxylating alpha subunit ISP domain
63 d1bxba_
not modelled
10.9
10
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: Xylose isomerase
64 c3bjxB_
not modelled
10.3
14
PDB header: hydrolaseChain: B: PDB Molecule: halocarboxylic acid dehalogenase dehi;PDBTitle: structure of a group i haloacid dehalogenase from2 pseudomonas putida strain pp3
65 c4gq6A_
not modelled
10.2
32
PDB header: transcription/transcription inhibitorChain: A: PDB Molecule: menin;PDBTitle: human menin in complex with mll peptide
66 c2mkvA_
not modelled
10.1
24
PDB header: transport proteinChain: A: PDB Molecule: sodium/potassium-transporting atpase subunit gamma;PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
67 d1ygya4
not modelled
10.1
19
Fold: FwdE/GAPDH domain-likeSuperfamily: Serine metabolism enzymes domainFamily: SerA intervening domain-like
68 c1rfoC_
not modelled
10.0
33
PDB header: viral proteinChain: C: PDB Molecule: whisker antigen control protein;PDBTitle: trimeric foldon of the t4 phagehead fibritin
69 c6hwhB_
not modelled
9.9
22
PDB header: electron transportChain: B: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
70 c5xtdo_
not modelled
9.8
10
PDB header: oxidoreductase/electron transportChain: O: PDB Molecule: nadh dehydrogenase [ubiquinone] flavoprotein 2,PDBTitle: cryo-em structure of human respiratory complex i
71 c2kvoA_
not modelled
9.7
13
PDB header: photosynthesisChain: A: PDB Molecule: photosystem ii reaction center psb28 protein;PDBTitle: solution nmr structure of photosystem ii reaction center psb28 protein2 from synechocystis sp.(strain pcc 6803), northeast structural3 genomics consortium target sgr171
72 c2l82A_
not modelled
9.5
13
PDB header: de novo proteinChain: A: PDB Molecule: designed protein or32;PDBTitle: solution nmr structure of de novo designed protein, p-loop ntpase2 fold, northeast structural genomics consortium target or32
73 c5ggoB_
not modelled
9.4
18
PDB header: sugar binding proteinChain: B: PDB Molecule: protein o-linked-mannose beta-1,2-n-PDBTitle: crystal structure of n-terminal domain of human protein o-mannose2 beta-1,2-n-acetylglucosaminyltransferase in complex with galnac-3 beta1,3-glcnac-beta-pnp
74 c5zihA_
not modelled
9.3
7
PDB header: membrane proteinChain: A: PDB Molecule: sensory opsin a,chrimson;PDBTitle: crystal structure of the red light-activated channelrhodopsin2 chrimson.
75 c3qxbB_
not modelled
9.0
6
PDB header: isomeraseChain: B: PDB Molecule: putative xylose isomerase;PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
76 c4v19U_
not modelled
8.7
25
PDB header: ribosomeChain: U: PDB Molecule: mitoribosomal protein bl20m, mrpl20;PDBTitle: structure of the large subunit of the mammalian mitoribosome, part 12 of 2
77 d2nn6c2
not modelled
8.4
16
Fold: Ribonuclease PH domain 2-likeSuperfamily: Ribonuclease PH domain 2-likeFamily: Ribonuclease PH domain 2-like
78 c5o60R_
not modelled
8.4
39
PDB header: ribosomeChain: R: PDB Molecule: 50s ribosomal protein l20;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
79 d2zjrn1
not modelled
8.3
39
Fold: PABP domain-likeSuperfamily: Ribosomal protein L20Family: Ribosomal protein L20
80 d1c4ka2
not modelled
8.3
24
Fold: PLP-dependent transferase-likeSuperfamily: PLP-dependent transferasesFamily: Ornithine decarboxylase major domain
81 c4hyjB_
not modelled
8.3
6
PDB header: proton transportChain: B: PDB Molecule: rhodopsin;PDBTitle: crystal structure of exiguobacterium sibiricum rhodopsin
82 c5mlc4_
not modelled
8.2
65
PDB header: ribosomeChain: 4: PDB Molecule: 50s ribosomal protein l34, chloroplastic;PDBTitle: cryo-em structure of the spinach chloroplast ribosome reveals the2 location of plastid-specific ribosomal proteins and extensions
83 c6csmC_
not modelled
8.0
18
PDB header: membrane proteinChain: C: PDB Molecule: gtacr1;PDBTitle: crystal structure of the natural light-gated anion channel gtacr1
84 c5mc9C_
not modelled
7.9
19
PDB header: cell adhesionChain: C: PDB Molecule: laminin subunit gamma-1;PDBTitle: crystal structure of the heterotrimeric integrin-binding region of2 laminin-111
85 c3j3wQ_
not modelled
7.9
39
PDB header: ribosomeChain: Q: PDB Molecule: 50s ribosomal protein l20;PDBTitle: atomic model of the immature 50s subunit from bacillus subtilis (state2 ii-a)
86 c3wt9A_
not modelled
7.7
5
PDB header: proton transportChain: A: PDB Molecule: rhodopsin i;PDBTitle: crystal structure of the cell-free synthesized membrane protein,2 acetabularia rhodopsin i, at 1.48 angstrom
87 d2j01u1
not modelled
7.5
32
Fold: PABP domain-likeSuperfamily: Ribosomal protein L20Family: Ribosomal protein L20
88 c4hqjG_
not modelled
7.5
25
PDB header: hydrolase/transport proteinChain: G: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of na+,k+-atpase in the na+-bound state
89 c3am6C_
not modelled
7.3
10
PDB header: transport proteinChain: C: PDB Molecule: rhodopsin-2;PDBTitle: crystal structure of the proton pumping rhodopsin ar2 from marine alga2 acetabularia acetabulum
90 c6q8iK_
not modelled
7.2
27
PDB header: splicingChain: K: PDB Molecule: protein red;PDBTitle: nterminal domain of human smu1 in complex with human redmid
91 d2gtaa1
not modelled
6.9
9
Fold: all-alpha NTP pyrophosphatasesSuperfamily: all-alpha NTP pyrophosphatasesFamily: MazG-like
92 d2iuba2
not modelled
6.8
8
Fold: Transmembrane helix hairpinSuperfamily: Magnesium transport protein CorA, transmembrane regionFamily: Magnesium transport protein CorA, transmembrane region
93 c2e1cA_
not modelled
6.8
8
PDB header: transcription/dnaChain: A: PDB Molecule: putative hth-type transcriptional regulator ph1519;PDBTitle: structure of putative hth-type transcriptional regulator ph1519/dna2 complex
94 d1eb7a2
not modelled
6.7
14
Fold: Cytochrome cSuperfamily: Cytochrome cFamily: Di-heme cytochrome c peroxidase
95 c4hqjE_
not modelled
6.6
25
PDB header: hydrolase/transport proteinChain: E: PDB Molecule: na+/k+ atpase gamma subunit transcript variant a;PDBTitle: crystal structure of na+,k+-atpase in the na+-bound state
96 c4eb5B_
not modelled
6.5
19
PDB header: transferase/metal binding proteinChain: B: PDB Molecule: probable cysteine desulfurase 2;PDBTitle: a. fulgidus iscs-iscu complex structure
97 c3a24A_
not modelled
6.4
16
PDB header: hydrolaseChain: A: PDB Molecule: alpha-galactosidase;PDBTitle: crystal structure of bt1871 retaining glycosidase
98 c2ostC_
not modelled
6.3
19
PDB header: hydrolase/dnaChain: C: PDB Molecule: putative endonuclease;PDBTitle: the structure of a bacterial homing endonuclease : i-ssp6803i
99 c3d0fA_
not modelled
6.3
20
PDB header: transferaseChain: A: PDB Molecule: penicillin-binding 1 transmembrane protein mrca;PDBTitle: structure of the big_1156.2 domain of putative penicillin-binding2 protein mrca from nitrosomonas europaea atcc 19718