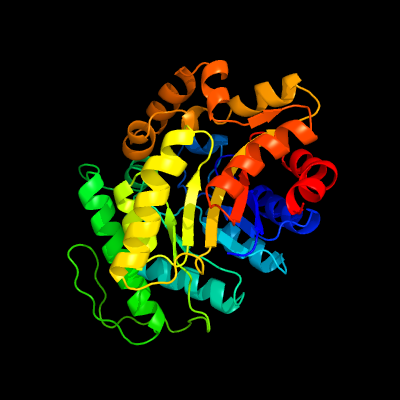
| 1 | c3c8nB_
|
|
 |
100.0 |
97 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable f420-dependent glucose-6-phosphate dehydrogenase
PDBTitle: crystal structure of apo-fgd1 from mycobacterium tuberculosis
|
|
|
|
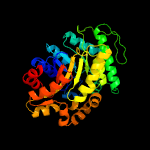
| 2 | d1rhca_
|
|
 |
100.0 |
34 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
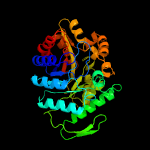
| 3 | d1ezwa_
|
|
 |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
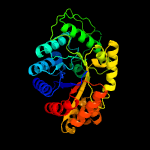
| 4 | d1luca_
|
|
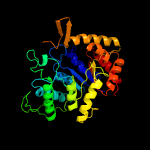 |
100.0 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
| 5 | c1tvlA_
|
|
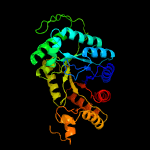 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:protein ytnj;
PDBTitle: structure of ytnj from bacillus subtilis
|
|
|
|
| 6 | d1tvla_
|
|
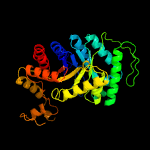 |
100.0 |
16 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 7 | c5tlcA_
|
|
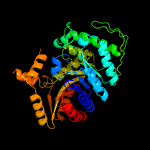 |
100.0 |
20 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dibenzothiophene desulfurization enzyme a;
PDBTitle: crystal structure of bdsa from bacillus subtilis wu-s2b
|
|
|
|
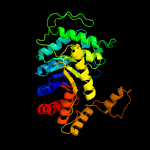
| 8 | c3sdoB_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nitrilotriacetate monooxygenase;
PDBTitle: structure of a nitrilotriacetate monooxygenase from burkholderia2 pseudomallei
|
|
|
|
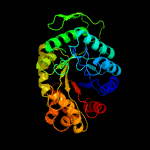
| 9 | c1z69D_
|
|
 |
100.0 |
22 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:coenzyme f420-dependent n(5),n(10)-
PDBTitle: crystal structure of methylenetetrahydromethanopterin2 reductase (mer) in complex with coenzyme f420
|
|
|
|
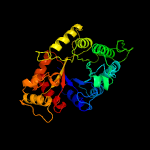
| 10 | c3raoB_
|
|
 |
100.0 |
17 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus atcc 10987.
|
|
|
|
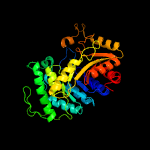
| 11 | c5wanA_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:pyrimidine monooxygenase ruta;
PDBTitle: crystal structure of a flavoenzyme ruta in the pyrimidine catabolic2 pathway
|
|
|
|
| 12 | c5w4zA_
|
|
 |
100.0 |
20 |
PDB header:flavoprotein
Chain: A: PDB Molecule:riboflavin lyase;
PDBTitle: crystal structure of riboflavin lyase (rcae) with modified fmn and2 substrate riboflavin
|
|
|
|
| 13 | d1lucb_
|
|
 |
100.0 |
13 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Bacterial luciferase (alkanal monooxygenase) |
|
|
|
| 14 | c2i7gA_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:monooxygenase;
PDBTitle: crystal structure of monooxygenase from agrobacterium tumefaciens
|
|
|
|
| 15 | c3b9nB_
|
|
 |
100.0 |
16 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:alkane monoxygenase;
PDBTitle: crystal structure of long-chain alkane monooxygenase (lada)
|
|
|
|
| 16 | c5dqpA_
|
|
 |
100.0 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:edta monooxygenase;
PDBTitle: edta monooxygenase (emoa) from chelativorans sp. bnc1
|
|
|
|
| 17 | c2wgkA_
|
|
 |
100.0 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3,6-diketocamphane 1,6 monooxygenase;
PDBTitle: type ii baeyer-villiger monooxygenase oxygenating2 constituent of 3,6-diketocamphane 1,6 monooxygenase from3 pseudomonas putida
|
|
|
|
| 18 | d1f07a_
|
|
 |
100.0 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:F420 dependent oxidoreductases |
|
|
|
| 19 | d1nqka_
|
|
 |
100.0 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Ssud-like monoxygenases |
|
|
|
| 20 | c6friD_
|
|
 |
100.0 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alkanal monooxygenase beta chain;
PDBTitle: structure of luxb from photobacterium leiognathi
|
|
|
|
| 21 | c6ak1B_ |
|
not modelled |
100.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:dimethyl-sulfide monooxygenase;
PDBTitle: crystal structure of dmoa from hyphomicrobium sulfonivorans
|
|
|
| 22 | c2b81D_ |
|
not modelled |
100.0 |
20 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:luciferase-like monooxygenase;
PDBTitle: crystal structure of the luciferase-like monooxygenase from bacillus2 cereus
|
|
|
| 23 | d1nfpa_ |
|
not modelled |
100.0 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 24 | d1fvpa_ |
|
not modelled |
99.5 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:Bacterial luciferase-like
Family:Non-fluorescent flavoprotein (luxF, FP390) |
|
|
| 25 | d1jpdx1 |
|
not modelled |
75.6 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 26 | c5zmyF_ |
|
not modelled |
74.9 |
22 |
PDB header:hydrolase
Chain: F: PDB Molecule:cis-epoxysuccinate hydrolase;
PDBTitle: crystal structure of a cis-epoxysuccinate hydrolase producing d(-)-2 tartaric acids
|
|
|
| 27 | c3qy6A_ |
|
not modelled |
74.5 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase ywqe;
PDBTitle: crystal structures of ywqe from bacillus subtilis and cpsb from2 streptococcus pneumoniae, unique metal-dependent tyrosine3 phosphatases
|
|
|
| 28 | c4nncA_ |
|
not modelled |
65.9 |
17 |
PDB header:lyase
Chain: A: PDB Molecule:obca, oxalate biosynthetic component a;
PDBTitle: ternary complex of obca with c4-coa adduct and oxalate
|
|
|
| 29 | c2uvaI_ |
|
not modelled |
65.1 |
19 |
PDB header:transferase
Chain: I: PDB Molecule:fatty acid synthase beta subunits;
PDBTitle: crystal structure of fatty acid synthase from thermomyces2 lanuginosus at 3.1 angstrom resolution. this file contains3 the beta subunits of the fatty acid synthase. the entire4 crystal structure consists of one heterododecameric fatty5 acid synthase and is described in remark 400
|
|
|
| 30 | c2x7vA_ |
|
not modelled |
64.5 |
10 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable endonuclease 4;
PDBTitle: crystal structure of thermotoga maritima endonuclease iv in2 the presence of zinc
|
|
|
| 31 | d1jpma1 |
|
not modelled |
63.5 |
18 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 32 | c4b3yB_ |
|
not modelled |
62.4 |
25 |
PDB header:transferase
Chain: B: PDB Molecule:fatty acid synthase;
PDBTitle: cryo-em structure of the mycobacterial fatty acid synthase
|
|
|
| 33 | c3b8iF_ |
|
not modelled |
61.1 |
17 |
PDB header:lyase
Chain: F: PDB Molecule:pa4872 oxaloacetate decarboxylase;
PDBTitle: crystal structure of oxaloacetate decarboxylase from pseudomonas2 aeruginosa (pa4872) in complex with oxalate and mg2+.
|
|
|
| 34 | c3chvA_ |
|
not modelled |
57.1 |
17 |
PDB header:metal binding protein
Chain: A: PDB Molecule:prokaryotic domain of unknown function (duf849) with a tim
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 member (spoa0042) from silicibacter pomeroyi dss-3 at 1.45 a3 resolution
|
|
|
| 35 | c3fa4D_ |
|
not modelled |
56.4 |
18 |
PDB header:lyase
Chain: D: PDB Molecule:2,3-dimethylmalate lyase;
PDBTitle: crystal structure of 2,3-dimethylmalate lyase, a pep mutase/isocitrate2 lyase superfamily member, triclinic crystal form
|
|
|
| 36 | d2d69a1 |
|
not modelled |
53.1 |
9 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 37 | c3no5C_ |
|
not modelled |
52.7 |
15 |
PDB header:structural genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a pfam duf849 domain containing protein2 (reut_a1631) from ralstonia eutropha jmp134 at 1.90 a resolution
|
|
|
| 38 | c2vkzH_ |
|
not modelled |
52.6 |
15 |
PDB header:transferase
Chain: H: PDB Molecule:fatty acid synthase subunit beta;
PDBTitle: structure of the cerulenin-inhibited fungal fatty acid synthase type i2 multienzyme complex
|
|
|
| 39 | d1muma_ |
|
not modelled |
52.5 |
22 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 40 | c3eooL_ |
|
not modelled |
52.2 |
19 |
PDB header:lyase
Chain: L: PDB Molecule:methylisocitrate lyase;
PDBTitle: 2.9a crystal structure of methyl-isocitrate lyase from2 burkholderia pseudomallei
|
|
|
| 41 | c3wqoB_ |
|
not modelled |
52.2 |
19 |
PDB header:unknown function
Chain: B: PDB Molecule:uncharacterized protein mj1311;
PDBTitle: crystal structure of d-tagatose 3-epimerase-like protein
|
|
|
| 42 | c1zlpA_ |
|
not modelled |
51.5 |
11 |
PDB header:lyase
Chain: A: PDB Molecule:petal death protein;
PDBTitle: petal death protein psr132 with cysteine-linked glutaraldehyde forming2 a thiohemiacetal adduct
|
|
|
| 43 | d1ujqa_ |
|
not modelled |
50.9 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 44 | c3e02A_ |
|
not modelled |
50.3 |
24 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849;
PDBTitle: crystal structure of a duf849 family protein (bxe_c0271) from2 burkholderia xenovorans lb400 at 1.90 a resolution
|
|
|
| 45 | c2ze3A_ |
|
not modelled |
48.8 |
19 |
PDB header:isomerase
Chain: A: PDB Molecule:dfa0005;
PDBTitle: crystal structure of dfa0005 complexed with alpha-ketoglutarate: a2 novel member of the icl/pepm superfamily from alkali-tolerant3 deinococcus ficus
|
|
|
| 46 | c3lyeA_ |
|
not modelled |
47.9 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:oxaloacetate acetyl hydrolase;
PDBTitle: crystal structure of oxaloacetate acetylhydrolase
|
|
|
| 47 | c2qiwA_ |
|
not modelled |
47.5 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:pep phosphonomutase;
PDBTitle: crystal structure of a putative phosphoenolpyruvate phosphonomutase2 (ncgl1015, cgl1060) from corynebacterium glutamicum atcc 13032 at3 1.80 a resolution
|
|
|
| 48 | c3c6cA_ |
|
not modelled |
45.6 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of a putative 3-keto-5-aminohexanoate cleavage2 enzyme (reut_c6226) from ralstonia eutropha jmp134 at 1.72 a3 resolution
|
|
|
| 49 | c2wjeA_ |
|
not modelled |
44.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:tyrosine-protein phosphatase cpsb;
PDBTitle: crystal structure of the tyrosine phosphatase cps4b from2 steptococcus pneumoniae tigr4.
|
|
|
| 50 | d2dfaa1 |
|
not modelled |
44.0 |
19 |
Fold:7-stranded beta/alpha barrel
Superfamily:Glycoside hydrolase/deacetylase
Family:LamB/YcsF-like |
|
|
| 51 | d1tzza1 |
|
not modelled |
43.7 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:Enolase C-terminal domain-like
Family:D-glucarate dehydratase-like |
|
|
| 52 | c5ud6B_ |
|
not modelled |
43.5 |
13 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dhdps from cyanidioschyzon merolae with lysine2 bound
|
|
|
| 53 | c2y7eA_ |
|
not modelled |
42.7 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:3-keto-5-aminohexanoate cleavage enzyme;
PDBTitle: crystal structure of the 3-keto-5-aminohexanoate cleavage enzyme2 (kce) from candidatus cloacamonas acidaminovorans (tetragonal form)
|
|
|
| 54 | c3ih1A_ |
|
not modelled |
41.0 |
26 |
PDB header:lyase
Chain: A: PDB Molecule:methylisocitrate lyase;
PDBTitle: crystal structure of carboxyvinyl-carboxyphosphonate phosphorylmutase2 from bacillus anthracis
|
|
|
| 55 | d1geha1 |
|
not modelled |
40.5 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 56 | c2d69B_ |
|
not modelled |
40.1 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: crystal structure of the complex of sulfate ion and octameric2 ribulose-1,5-bisphosphate carboxylase/oxygenase (rubisco) from3 pyrococcus horikoshii ot3 (form-2 crystal)
|
|
|
| 57 | d1s2wa_ |
|
not modelled |
39.8 |
23 |
Fold:TIM beta/alpha-barrel
Superfamily:Phosphoenolpyruvate/pyruvate domain
Family:Phosphoenolpyruvate mutase/Isocitrate lyase-like |
|
|
| 58 | c4mwaA_ |
|
not modelled |
37.9 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: 1.85 angstrom crystal structure of gcpe protein from bacillus2 anthracis
|
|
|
| 59 | c1rldB_ |
|
not modelled |
37.8 |
12 |
PDB header:lyase(carbon-carbon)
Chain: B: PDB Molecule:ribulose 1,5 bisphosphate carboxylase/oxygenase (large
PDBTitle: solid-state phase transition in the crystal structure of ribulose 1,5-2 biphosphate carboxylase(slash)oxygenase
|
|
|
| 60 | c6hunA_ |
|
not modelled |
36.4 |
9 |
PDB header:photosynthesis
Chain: A: PDB Molecule:ribulose bisphosphate carboxylase;
PDBTitle: dimeric archeal rubisco from hyperthermus butylicus
|
|
|
| 61 | c6daoB_ |
|
not modelled |
36.4 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:trans-o-hydroxybenzylidenepyruvate hydratase-aldolase;
PDBTitle: nahe wt selenomethionine
|
|
|
| 62 | c3umuA_ |
|
not modelled |
34.7 |
21 |
PDB header:hydrolase
Chain: A: PDB Molecule:histidinol-phosphatase;
PDBTitle: crystal structure of l-histidinol phosphate phosphatase (hisk) from2 lactococcus lactis subsp. lactis il1403 complexed with zn, phosphate3 and l-histidinol
|
|
|
| 63 | c2r8wB_ |
|
not modelled |
33.9 |
16 |
PDB header:lyase
Chain: B: PDB Molecule:agr_c_1641p;
PDBTitle: the crystal structure of dihydrodipicolinate synthase (atu0899) from2 agrobacterium tumefaciens str. c58
|
|
|
| 64 | c4rtbA_ |
|
not modelled |
33.7 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:hydg protein;
PDBTitle: x-ray structure of the fefe-hydrogenase maturase hydg from2 carboxydothermus hydrogenoformans
|
|
|
| 65 | c4qccA_ |
|
not modelled |
33.7 |
18 |
PDB header:structural protein, lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase, peptidyl-
PDBTitle: structure of a cube-shaped, highly porous protein cage designed by2 fusing symmetric oligomeric domains
|
|
|
| 66 | c4mg4G_ |
|
not modelled |
33.5 |
17 |
PDB header:unknown function
Chain: G: PDB Molecule:phosphonomutase;
PDBTitle: crystal structure of a putative phosphonomutase from burkholderia2 cenocepacia j2315
|
|
|
| 67 | c4lsbA_ |
|
not modelled |
31.9 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:lyase/mutase;
PDBTitle: crystal structure of a putative lyase/mutase from burkholderia2 cenocepacia j2315
|
|
|
| 68 | c1r30A_ |
|
not modelled |
31.7 |
8 |
PDB header:transferase
Chain: A: PDB Molecule:biotin synthase;
PDBTitle: the crystal structure of biotin synthase, an s-adenosylmethionine-2 dependent radical enzyme
|
|
|
| 69 | d1r30a_ |
|
not modelled |
31.7 |
8 |
Fold:TIM beta/alpha-barrel
Superfamily:Radical SAM enzymes
Family:Biotin synthase |
|
|
| 70 | c1rcxH_ |
|
not modelled |
30.4 |
12 |
PDB header:lyase (carbon-carbon)
Chain: H: PDB Molecule:ribulose bisphosphate carboxylase/oxygenase;
PDBTitle: non-activated spinach rubisco in complex with its substrate2 ribulose-1,5-bisphosphate
|
|
|
| 71 | d1o5ka_ |
|
not modelled |
30.3 |
17 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 72 | c1gehE_ |
|
not modelled |
30.2 |
19 |
PDB header:lyase
Chain: E: PDB Molecule:ribulose-1,5-bisphosphate carboxylase/oxygenase;
PDBTitle: crystal structure of archaeal rubisco (ribulose 1,5-bisphosphate2 carboxylase/oxygenase)
|
|
|
| 73 | c3lotC_ |
|
not modelled |
29.1 |
20 |
PDB header:structure genomics, unknown function
Chain: C: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of protein of unknown function (np_070038.1) from2 archaeoglobus fulgidus at 1.89 a resolution
|
|
|
| 74 | c6daqA_ |
|
not modelled |
28.0 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:phdj;
PDBTitle: phdj bound to substrate intermediate
|
|
|
| 75 | c5k9xA_ |
|
not modelled |
27.9 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha chain from legionella2 pneumophila subsp. pneumophila
|
|
|
| 76 | d8ruca1 |
|
not modelled |
27.0 |
10 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 77 | c4wd0A_ |
|
not modelled |
26.1 |
21 |
PDB header:isomerase
Chain: A: PDB Molecule:1-(5-phosphoribosyl)-5-[(5-phosphoribosylamino)
PDBTitle: crystal structure of hisap form arthrobacter aurescens
|
|
|
| 78 | c5kinC_ |
|
not modelled |
25.8 |
24 |
PDB header:lyase
Chain: C: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase alpha beta complex from2 streptococcus pneumoniae
|
|
|
| 79 | d1ykwa1 |
|
not modelled |
25.2 |
31 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 80 | c3noyA_ |
|
not modelled |
24.8 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-en-1-yl diphosphate synthase;
PDBTitle: crystal structure of ispg (gcpe)
|
|
|
| 81 | c3pueA_ |
|
not modelled |
24.5 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of the complex of dhydrodipicolinate synthase from2 acinetobacter baumannii with lysine at 2.6a resolution
|
|
|
| 82 | c1bf2A_ |
|
not modelled |
23.8 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:isoamylase;
PDBTitle: structure of pseudomonas isoamylase
|
|
|
| 83 | c2v82A_ |
|
not modelled |
23.3 |
18 |
PDB header:lyase
Chain: A: PDB Molecule:2-dehydro-3-deoxy-6-phosphogalactonate aldolase;
PDBTitle: kdpgal complexed to kdpgal
|
|
|
| 84 | d1wdda1 |
|
not modelled |
23.3 |
12 |
Fold:TIM beta/alpha-barrel
Superfamily:RuBisCo, C-terminal domain
Family:RuBisCo, large subunit, C-terminal domain |
|
|
| 85 | c3d0cB_ |
|
not modelled |
23.0 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from oceanobacillus2 iheyensis at 1.9 a resolution
|
|
|
| 86 | c1zfjA_ |
|
not modelled |
22.7 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:inosine monophosphate dehydrogenase;
PDBTitle: inosine monophosphate dehydrogenase (impdh; ec 1.1.1.205) from2 streptococcus pyogenes
|
|
|
| 87 | c3e49A_ |
|
not modelled |
22.6 |
22 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein duf849 with a tim barrel fold;
PDBTitle: crystal structure of a prokaryotic domain of unknown function (duf849)2 with a tim barrel fold (bxe_c0966) from burkholderia xenovorans lb4003 at 1.75 a resolution
|
|
|
| 88 | d2a6na1 |
|
not modelled |
22.3 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 89 | c3qfeB_ |
|
not modelled |
22.1 |
14 |
PDB header:lyase
Chain: B: PDB Molecule:putative dihydrodipicolinate synthase family protein;
PDBTitle: crystal structures of a putative dihydrodipicolinate synthase family2 protein from coccidioides immitis
|
|
|
| 90 | c6mywA_ |
|
not modelled |
22.0 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:n-ethylmaleimide reductase;
PDBTitle: gluconobacter ene-reductase (gluer) mutant - t36a
|
|
|
| 91 | d1xxxa1 |
|
not modelled |
22.0 |
15 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 92 | d1xcfa_ |
|
not modelled |
21.9 |
11 |
Fold:TIM beta/alpha-barrel
Superfamily:Ribulose-phoshate binding barrel
Family:Tryptophan biosynthesis enzymes |
|
|
| 93 | c3lerA_ |
|
not modelled |
21.7 |
19 |
PDB header:lyase
Chain: A: PDB Molecule:dihydrodipicolinate synthase;
PDBTitle: crystal structure of dihydrodipicolinate synthase from campylobacter2 jejuni subsp. jejuni nctc 11168
|
|
|
| 94 | c4a3uB_ |
|
not modelled |
21.4 |
13 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadh\:flavin oxidoreductase/nadh oxidase;
PDBTitle: x-structure of the old yellow enzyme homologue from zymomonas mobilis2 (ncr)
|
|
|
| 95 | d1ht6a2 |
|
not modelled |
21.4 |
19 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Amylase, catalytic domain |
|
|
| 96 | d1f74a_ |
|
not modelled |
20.9 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|
| 97 | d1x7fa2 |
|
not modelled |
20.8 |
24 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Outer surface protein, N-terminal domain |
|
|
| 98 | c4qnwA_ |
|
not modelled |
20.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:chanoclavine-i aldehyde reductase;
PDBTitle: crystal structure of easa, an old yellow enzyme from aspergillus2 fumigatus
|
|
|
| 99 | c2p0oA_ |
|
not modelled |
20.5 |
15 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein duf871;
PDBTitle: crystal structure of a conserved protein from locus ef_2437 in2 enterococcus faecalis with an unknown function
|
|
|
| 100 | d1wa3a1 |
|
not modelled |
20.5 |
14 |
Fold:TIM beta/alpha-barrel
Superfamily:Aldolase
Family:Class I aldolase |
|
|