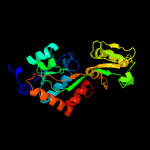
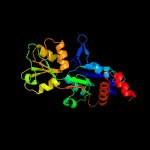
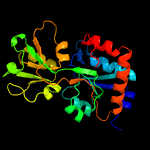
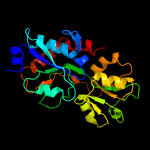
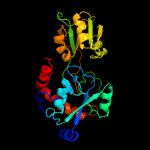
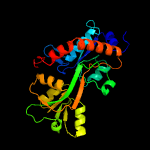
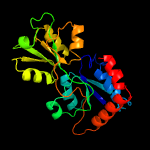
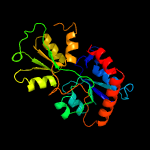
1 c6h2tA_
100.0
100
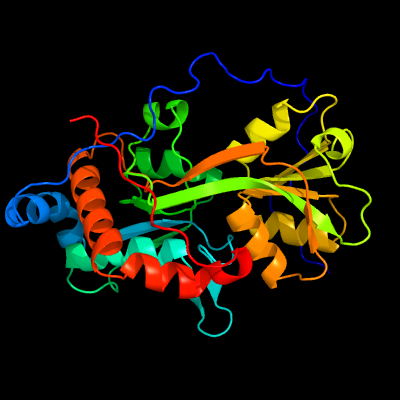
PDB header: signaling proteinChain: A: PDB Molecule: probable glutamine-binding lipoprotein glnh (glnbp);PDBTitle: glnh bound to glu, mycobacterium tuberculosis
2 c4z9nB_
100.0
28
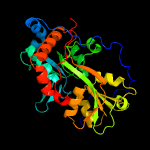
PDB header: transport proteinChain: B: PDB Molecule: amino acid abc transporter, periplasmic amino acid-bindingPDBTitle: abc transporter / periplasmic binding protein from brucella ovis with2 glutathione bound
3 c5eyfA_
100.0
33
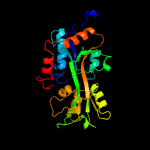
PDB header: solute-binding proteinChain: A: PDB Molecule: glutamate abc superfamily atp binding cassette transporter,PDBTitle: crystal structure of solute-binding protein from enterococcus faecium2 with bound glutamate
4 c4h5fB_
100.0
21
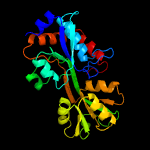
PDB header: transport proteinChain: B: PDB Molecule: amino acid abc superfamily atp binding cassettePDBTitle: crystal structure of an amino acid abc transporter substrate-binding2 protein from streptococcus pneumoniae canada mdr_19a bound to l-3 arginine, form 1
5 c3vvfA_
100.0
22
PDB header: transport proteinChain: A: PDB Molecule: amino acid abc transporter, binding protein;PDBTitle: crystal structure of ttc0807 complexed with arginine
6 c6detA_
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: tv2483;PDBTitle: the crystal structure of tv2483 bound to l-arginine
7 c2ylnA_
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: putative abc transporter, periplasmic binding protein,PDBTitle: crystal structure of the l-cystine solute receptor of2 neisseria gonorrhoeae in the closed conformation
8 c3hv1A_
100.0
18
PDB header: transport proteinChain: A: PDB Molecule: polar amino acid abc uptake transporter substratePDBTitle: crystal structure of a polar amino acid abc uptake2 transporter substrate binding protein from streptococcus3 thermophilus
9 c4pshA_
100.0
28
PDB header: protein transportChain: A: PDB Molecule: abc-type transporter, periplasmic subunit family 3;PDBTitle: structure of holo argbp from t. maritima
10 c4ohnA_
100.0
21
PDB header: transport proteinChain: A: PDB Molecule: abc transporter substrate-binding protein;PDBTitle: crystal structure of an abc uptake transporter substrate binding2 protein from streptococcus pneumoniae with bound histidine
11 c4oz9A_
100.0
19
PDB header: lyaseChain: A: PDB Molecule: membrane-bound lytic murein transglycosylase f;PDBTitle: crystal structure of mltf from pseudomonas aeruginosa complexed with2 isoleucine
12 c4q0cA_
100.0
19
PDB header: transferaseChain: A: PDB Molecule: virulence sensor protein bvgs;PDBTitle: 3.1 a resolution crystal structure of the b. pertussis bvgs2 periplasmic domain
13 c2vhaB_
100.0
22
PDB header: transport proteinChain: B: PDB Molecule: periplasmic binding transport protein;PDBTitle: debp
14 c3kzgB_
100.0
26
PDB header: transport proteinChain: B: PDB Molecule: arginine 3rd transport system periplasmic bindingPDBTitle: crystal structure of an arginine 3rd transport system2 periplasmic binding protein from legionella pneumophila
15 c4pp0B_
100.0
21
PDB header: transport proteinChain: B: PDB Molecule: nopaline-binding periplasmic protein;PDBTitle: structure of the pbp noct-m117n in complex with pyronopaline
16 c3k4uA_
100.0
22
PDB header: transport proteinChain: A: PDB Molecule: binding component of abc transporter;PDBTitle: crystal structure of putative binding component of abc transporter2 from wolinella succinogenes dsm 1740 complexed with lysine
17 c5hmtA_
100.0
17
PDB header: transport proteinChain: A: PDB Molecule: cyclohexadienyl dehydratase;PDBTitle: crystal structure of the cyclohexadienyl dehydratase-like solute-2 binding protein sar11_1068 from candidatus pelagibacter ubique.
18 d2a5sa1
100.0
20
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
19 c5josA_
100.0
25
PDB header: lyaseChain: A: PDB Molecule: cyclohexadienyl dehydratase;PDBTitle: crystal structure of an ancestral cyclohexadienyl dehydratase, anccdt-2 3(p188l).
20 c1xt8B_
100.0
22
PDB header: transport proteinChain: B: PDB Molecule: putative amino-acid transporter periplasmic solute-bindingPDBTitle: crystal structure of cysteine-binding protein from campylobacter2 jejuni at 2.0 a resolution
21 c5kuhB_
not modelled
100.0
18
PDB header: signaling proteinChain: B: PDB Molecule: glutamate receptor ionotropic, kainate 2;PDBTitle: gluk2em with ly466195
22 c2v25B_
not modelled
100.0
27
PDB header: receptorChain: B: PDB Molecule: major cell-binding factor;PDBTitle: structure of the campylobacter jejuni antigen peb1a, an2 aspartate and glutamate receptor with bound aspartate
23 c2yjpB_
not modelled
100.0
26
PDB header: transport proteinChain: B: PDB Molecule: putative abc transporter, periplasmic binding protein,PDBTitle: crystal structure of the solute receptors for l-cysteine of2 neisseria gonorrhoeae
24 c2ieeB_
not modelled
100.0
18
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: probable abc transporter extracellular-binding proteinPDBTitle: crystal structure of yckb_bacsu from bacillus subtilis. northeast2 structural genomics consortium target sr574.
25 c4i62A_
not modelled
100.0
22
PDB header: transport proteinChain: A: PDB Molecule: amino acid abc transporter, periplasmic amino acid-bindingPDBTitle: 1.05 angstrom crystal structure of an amino acid abc transporter2 substrate-binding protein abpa from streptococcus pneumoniae canada3 mdr_19a bound to l-arginine
26 d1pb7a_
not modelled
100.0
17
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
27 c3kbrA_
not modelled
100.0
21
PDB header: lyaseChain: A: PDB Molecule: cyclohexadienyl dehydratase;PDBTitle: the crystal structure of cyclohexadienyl dehydratase precursor from2 pseudomonas aeruginosa pa01
28 c5orgA_
not modelled
100.0
22
PDB header: octopine-binding proteinChain: A: PDB Molecule: octopine-binding periplasmic protein;PDBTitle: structure of the periplasmic binding protein (pbp) occj from a.2 tumefaciens b6 in complex with octopine.
29 c5t0wA_
not modelled
100.0
33
PDB header: transport proteinChain: A: PDB Molecule: anccdt-1;PDBTitle: crystal structure of the ancestral amino acid-binding protein anccdt-2 1, a precursor of cyclohexadienyl dehydratase
30 c3g41A_
not modelled
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: amino acid abc transporter, periplasmic amino acid-bindingPDBTitle: the structure of cpn0482, the arginine binding protein from the2 periplasm of chlamydia pneumoniae
31 d1xt8a1
not modelled
100.0
22
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
32 c5uh0A_
not modelled
100.0
19
PDB header: hydrolase,oxidoreductaseChain: A: PDB Molecule: membrane-bound lytic murein transglycosylase f;PDBTitle: 1.95 angstrom resolution crystal structure of fragment (35-274) of2 membrane-bound lytic murein transglycosylase f from yersinia pestis.
33 c4ymxB_
not modelled
100.0
29
PDB header: transport proteinChain: B: PDB Molecule: abc-type amino acid transport system, periplasmicPDBTitle: crystal structure of the substrate binding protein of an amino acid2 abc transporter
34 c2q89A_
not modelled
100.0
16
PDB header: transport proteinChain: A: PDB Molecule: putative abc transporter amino acid-binding protein;PDBTitle: crystal structure of ehub in complex with hydroxyectoine
35 c3r39A_
not modelled
100.0
14
PDB header: transport proteinChain: A: PDB Molecule: putative periplasmic binding protein;PDBTitle: crystal structure of periplasmic d-alanine abc transporter from2 salmonella enterica
36 c2o1mB_
not modelled
100.0
21
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: probable amino-acid abc transporter extracellular-bindingPDBTitle: crystal structure of the probable amino-acid abc transporter2 extracellular-binding protein ytmk from bacillus subtilis. northeast3 structural genomics consortium target sr572
37 c2y7iB_
not modelled
100.0
22
PDB header: arginine-binding proteinChain: B: PDB Molecule: stm4351;PDBTitle: structural basis for high arginine specificity in salmonella2 typhimurium periplasmic binding protein stm4351.
38 c3delC_
not modelled
100.0
21
PDB header: protein binding, transport proteinChain: C: PDB Molecule: arginine binding protein;PDBTitle: the structure of ct381, the arginine binding protein from the2 periplasm chlamydia trachomatis
39 c4zv2A_
not modelled
100.0
32
PDB header: solute-binding proteinChain: A: PDB Molecule: ancqr;PDBTitle: an ancestral arginine-binding protein bound to glutamine
40 c2rc9A_
not modelled
100.0
22
PDB header: membrane proteinChain: A: PDB Molecule: glutamate [nmda] receptor subunit 3a;PDBTitle: crystal structure of the nr3a ligand binding core complex with acpc at2 1.96 angstrom resolution
41 c3i6vA_
not modelled
100.0
20
PDB header: transport proteinChain: A: PDB Molecule: periplasmic his/glu/gln/arg/opine family-binding protein;PDBTitle: crystal structure of a periplasmic his/glu/gln/arg/opine family-2 binding protein from silicibacter pomeroyi in complex with lysine
42 c3h7mA_
not modelled
100.0
21
PDB header: transferaseChain: A: PDB Molecule: sensor protein;PDBTitle: crystal structure of a histidine kinase sensor domain with similarity2 to periplasmic binding proteins
43 c5l2eB_
not modelled
100.0
19
PDB header: protein bindingChain: B: PDB Molecule: glutamate receptor ionotropic, delta-2,glutamate receptorPDBTitle: crystal structure of rat glutamate receptor delta-2 extracellular2 domain
44 c4oenA_
not modelled
100.0
24
PDB header: transport proteinChain: A: PDB Molecule: second substrate binding domain of putative amino acid abcPDBTitle: crystal structure of the second substrate binding domain of a putative2 amino acid abc transporter from streptococcus pneumoniae canada3 mdr_19a
45 c6a80B_
not modelled
100.0
23
PDB header: transport proteinChain: B: PDB Molecule: putative amino acid-binding periplasmic abc transporterPDBTitle: crystal structure of putative amino acid binding periplasmic abc2 transporter protein from candidatus liberibacter asiaticus in complex3 with cystine
46 c4c0rB_
not modelled
100.0
20
PDB header: transport proteinChain: B: PDB Molecule: putative amino acid binding protein;PDBTitle: molecular and structural basis of glutathione import in2 gram-positive bacteria via gsht and the cystine abc3 importer tcybc of streptococcus mutans.
47 c4uqqD_
not modelled
100.0
16
PDB header: transport proteinChain: D: PDB Molecule: glutamate receptor ionotropic, kainate 2;PDBTitle: electron density map of gluk2 desensitized state in complex with 2s,2 4r-4-methylglutamate
48 c4kr5B_
not modelled
100.0
17
PDB header: transport proteinChain: B: PDB Molecule: glutamine abc transporter permease and substrate bindingPDBTitle: crystal structure of lactococcus lactis glnp substrate binding domain2 2 (sbd2) in open conformation
49 c4g4pA_
not modelled
100.0
26
PDB header: transport proteinChain: A: PDB Molecule: amino acid abc transporter, amino acid-binding/permeasePDBTitle: crystal structure of glutamine-binding protein from enterococcus2 faecalis at 1.5 a
50 c2q2aD_
not modelled
100.0
27
PDB header: transport proteinChain: D: PDB Molecule: artj;PDBTitle: crystal structures of the arginine-, lysine-, histidine-binding2 protein artj from the thermophilic bacterium geobacillus3 stearothermophilus
51 c4eq9A_
not modelled
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: abc transporter substrate-binding protein-amino acidPDBTitle: 1.4 angstrom crystal structure of abc transporter glutathione-binding2 protein gsht from streptococcus pneumoniae strain canada mdr_19a in3 complex with glutathione
52 d1wdna_
not modelled
100.0
27
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
53 d1lsta_
not modelled
100.0
20
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
54 d1hsla_
not modelled
100.0
24
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
55 c5l9mA_
not modelled
100.0
20
PDB header: transport proteinChain: A: PDB Molecule: deoxyfructosyl-amino acid transporter periplasmic bindingPDBTitle: structure of agrobacterium tumefaciens b6 strain pbp soca complexed2 with deoxyfructosylglutamine (dfg)
56 c4f3pB_
not modelled
100.0
27
PDB header: transport proteinChain: B: PDB Molecule: glutamine-binding periplasmic protein;PDBTitle: crystal structure of a glutamine-binding periplasmic protein from2 burkholderia pseudomallei in complex with glutamine
57 c5dtbB_
not modelled
100.0
17
PDB header: membrane proteinChain: B: PDB Molecule: cg3822;PDBTitle: crystal structure of the drosophila cg3822 kair1d ligand binding2 domain complex with glutamate
58 c6mmvC_
not modelled
100.0
19
PDB header: transport proteinChain: C: PDB Molecule: glutamate receptor ionotropic, nmda 1;PDBTitle: triheteromeric nmda receptor glun1/glun2a/glun2a* extracellular domain2 in the '2-knuckle-asymmetric' conformation, in complex with glycine3 and glutamate, in the presence of 1 micromolar zinc chloride, and at4 ph 7.4
59 c4kptA_
not modelled
100.0
23
PDB header: transport proteinChain: A: PDB Molecule: glutamine abc transporter permease and substrate bindingPDBTitle: crystal structure of substrate binding domain 1 (sbd1) of abc2 transporter glnpq from lactococcus lactis
60 c3mplA_
not modelled
100.0
13
PDB header: signaling proteinChain: A: PDB Molecule: virulence sensor protein bvgs;PDBTitle: crystal structure of bordetella pertussis bvgs vft2 domain (double2 mutant f375e/q461e)
61 c5dt6A_
not modelled
100.0
18
PDB header: membrane proteinChain: A: PDB Molecule: glutamate receptor 1;PDBTitle: crystal structure of the drosophila glur1a ligand binding domain2 complex with glutamate
62 d1mqia_
not modelled
100.0
19
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
63 c2xx7B_
not modelled
100.0
19
PDB header: transport proteinChain: B: PDB Molecule: glutamate receptor 2;PDBTitle: crystal structure of 1-(4-(1-pyrrolidinylcarbonyl)phenyl)-3-2 (trifluoromethyl)-4,5,6,7-tetrahydro-1h-indazole in complex with3 the ligand binding domain of the rat glua2 receptor and glutamate4 at 2.2a resolution.
64 c4ykiA_
not modelled
100.0
15
PDB header: membrane proteinChain: A: PDB Molecule: ml032222a iglur;PDBTitle: mnemiopsis leidyi ml032222a iglur lbd glycine complex
65 c6mmvD_
not modelled
100.0
23
PDB header: transport proteinChain: D: PDB Molecule: glutamate receptor ionotropic, nmda 2a;PDBTitle: triheteromeric nmda receptor glun1/glun2a/glun2a* extracellular domain2 in the '2-knuckle-asymmetric' conformation, in complex with glycine3 and glutamate, in the presence of 1 micromolar zinc chloride, and at4 ph 7.4
66 c5iprD_
not modelled
100.0
25
PDB header: signaling proteinChain: D: PDB Molecule: ionotropic glutamate receptor subunit nr2b;PDBTitle: cryo-em structure of glun1/glun2b nmda receptor in the dcka/d-apv-2 bound conformation, state 3
67 c4zdmA_
not modelled
100.0
16
PDB header: membrane proteinChain: A: PDB Molecule: glutamate receptor kainate-like protein;PDBTitle: pleurobrachia bachei iglur3 lbd glycine complex
68 c5sv6A_
not modelled
100.0
16
PDB header: unknown functionChain: A: PDB Molecule: extracellular solute-binding protein, family 3;PDBTitle: crystal structure of mxaj from methlophaga aminisulfidivorans mpt
69 d1s50a1
not modelled
100.0
16
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
70 c4wxjB_
not modelled
100.0
17
PDB header: membrane proteinChain: B: PDB Molecule: glutamate receptor iib,glutamate receptor iib;PDBTitle: drosophila muscle gluriib complex with glutamate
71 c2pyyB_
not modelled
100.0
25
PDB header: transport proteinChain: B: PDB Molecule: ionotropic glutamate receptor bacterial homologue;PDBTitle: crystal structure of the glur0 ligand-binding core from nostoc2 punctiforme in complex with (l)-glutamate
72 d2f34a1
not modelled
100.0
15
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
73 c4io7A_
not modelled
100.0
18
PDB header: membrane proteinChain: A: PDB Molecule: avglur1 ligand binding domain;PDBTitle: crystal structure of the avglur1 ligand binding domain complex with2 phenylalanine at 1.9 angstrom resolution
74 d1ii5a_
not modelled
100.0
23
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
75 c5bwjC_
not modelled
100.0
15
PDB header: signaling proteinChain: C: PDB Molecule: sensory transduction histidine kinase, putative;PDBTitle: structural characterization and modeling of the borrelia burgdorferi2 hybrid histidine kinase hk1 periplasmic sensor
76 c1yaeB_
not modelled
100.0
16
PDB header: membrane proteinChain: B: PDB Molecule: glutamate receptor, ionotropic kainate 2;PDBTitle: structure of the kainate receptor subunit glur6 agonist binding domain2 complexed with domoic acid
77 c2v3tA_
not modelled
100.0
19
PDB header: receptorChain: A: PDB Molecule: glutamate receptor delta-2 subunit synonym glurdelta2, glurPDBTitle: structure of the ligand-binding core of the ionotropic glutamate2 receptor-like glurdelta2 in the apo form
78 c5ideA_
not modelled
100.0
21
PDB header: signaling proteinChain: A: PDB Molecule: glutamate receptor 2;PDBTitle: cryo-em structure of glua2/3 ampa receptor heterotetramer (model i)
79 c5ikbA_
not modelled
100.0
21
PDB header: membrane proteinChain: A: PDB Molecule: glutamate receptor ionotropic, kainate 4,glutamate receptorPDBTitle: crystal structure of the kainate receptor gluk4 ligand binding domain2 in complex with kainate
80 c6onpA_
not modelled
100.0
17
PDB header: unknown functionChain: A: PDB Molecule: periplasmic binding protein xoxj;PDBTitle: crystal structure of periplasmic binding protein xoxj from2 methylobacterium extorquens am1
81 c5kbuA_
not modelled
100.0
22
PDB header: transport proteinChain: A: PDB Molecule: glutamate receptor 2,voltage-dependent calcium channelPDBTitle: cryo-em structure of glua2-2xstz complex at 7.8 angstrom resolution
82 c4pe5A_
not modelled
100.0
19
PDB header: transport proteinChain: A: PDB Molecule: glutamate receptor ionotropic, nmda 1;PDBTitle: crystal structure of glun1a/glun2b nmda receptor ion channel
83 c4pe5B_
not modelled
100.0
19
PDB header: transport proteinChain: B: PDB Molecule: glutamate receptor ionotropic, nmda 2b;PDBTitle: crystal structure of glun1a/glun2b nmda receptor ion channel
84 c5gzsA_
not modelled
100.0
15
PDB header: signaling proteinChain: A: PDB Molecule: ggdef family protein;PDBTitle: structure of vc protein
85 c3kg2A_
not modelled
100.0
20
PDB header: membrane protein, transport proteinChain: A: PDB Molecule: glutamate receptor 2;PDBTitle: ampa subtype ionotropic glutamate receptor in complex with competitive2 antagonist zk 200775
86 c4tlmC_
not modelled
99.9
26
PDB header: signaling proteinChain: C: PDB Molecule: receptor subunit glun1;PDBTitle: crystal structure of glun1/glun2b nmda receptor, structure 2
87 c6mmiD_
not modelled
99.9
25
PDB header: transport proteinChain: D: PDB Molecule: glutamate receptor ionotropic, nmda 2a;PDBTitle: diheteromeric nmda receptor glun1/glun2a in the 'splayed-open'2 conformation, in complex with glycine and glutamate, in the presence3 of 1 millimolar zinc chloride, and at ph 7.4
88 c6qkzA_
not modelled
99.9
21
PDB header: membrane proteinChain: A: PDB Molecule: glua1;PDBTitle: full length glua1/2-gamma8 complex
89 c6irfD_
not modelled
99.9
21
PDB header: membrane proteinChain: D: PDB Molecule: glutamate receptor ionotropic, nmda 2a;PDBTitle: structure of the human glun1/glun2a nmda receptor in the2 glutamate/glycine-bound state at ph 6.3, class i
90 c6gpcB_
not modelled
99.9
33
PDB header: transport proteinChain: B: PDB Molecule: amino acid abc transporter, periplasmic amino acid-bindingPDBTitle: crystal structure of the arginine-bound form of domain 1 from tmargbp
91 c5uowB_
not modelled
99.9
24
PDB header: membrane proteinChain: B: PDB Molecule: n-methyl-d-aspartate receptor subunit nr2a;PDBTitle: triheteromeric nmda receptor glun1/glun2a/glun2b in complex with2 glycine, glutamate, mk-801 and a glun2b-specific fab, at ph 6.5
92 c5jvbB_
not modelled
99.8
11
PDB header: transport proteinChain: B: PDB Molecule: phosphonate abc transporter, periplasmic phosphonate-PDBTitle: 1.95a resolution structure of ptxb from trichodesmium erythraeum2 ims101 in complex with phosphite
93 c5o2kE_
not modelled
99.8
13
PDB header: transport proteinChain: E: PDB Molecule: probable phosphite transport system-binding protein ptxb;PDBTitle: native apo-structure of pseudomonas stutzeri ptxb to 2.1 a resolution
94 c5me4A_
not modelled
99.7
16
PDB header: transport proteinChain: A: PDB Molecule: probable phosphite transport system-binding protein htxb;PDBTitle: the structure of htxb from pseudomonas stutzeri in complex with2 hypophosphite to 1.52 a resolution
95 c5lv1C_
not modelled
99.7
12
PDB header: periplasmic binding proteinChain: C: PDB Molecule: ptxb;PDBTitle: 2.12 a resolution structure of ptxb from prochlorococcus marinus (mit2 9301) in complex with phosphite
96 c6esvA_
not modelled
99.5
13
PDB header: signaling proteinChain: A: PDB Molecule: putative periplasmic phosphite-binding-like protein (pbl)PDBTitle: structure of the phosphate-bound form of aiox from rhizobium sp. str.2 nt-26
97 c3n5lA_
not modelled
99.4
9
PDB header: transport proteinChain: A: PDB Molecule: binding protein component of abc phosphonate transporter;PDBTitle: crystal structure of a binding protein component of abc phosphonate2 transporter (pa3383) from pseudomonas aeruginosa at 1.97 a resolution
98 c3p7iA_
not modelled
99.4
12
PDB header: transport proteinChain: A: PDB Molecule: phnd, subunit of alkylphosphonate abc transporter;PDBTitle: crystal structure of escherichia coli phnd in complex with 2-2 aminoethyl phosphonate
99 c5ub6B_
not modelled
99.3
15
PDB header: metal binding proteinChain: B: PDB Molecule: phosphate-binding protein;PDBTitle: xac2383 from xanthomonas citri bound to pyrophosphate
100 c5lq8A_
not modelled
99.2
12
PDB header: periplasmic binding proteinChain: A: PDB Molecule: putative phosphonate binding protein for abc transporter;PDBTitle: 1.52 a resolution structure of phnd1 from prochlorococcus marinus (mit2 9301) in complex with methylphosphonate
101 c3e4rA_
not modelled
99.0
16
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein;PDBTitle: crystal structure of the alkanesulfonate binding protein2 (ssua) from the phytopathogenic bacteria xanthomonas3 axonopodis pv. citri bound to hepes
102 c2x26A_
not modelled
98.9
18
PDB header: transport proteinChain: A: PDB Molecule: periplasmic aliphatic sulphonates-binding protein;PDBTitle: crystal structure of the periplasmic aliphatic sulphonate binding2 protein ssua from escherichia coli
103 c3uifA_
not modelled
98.9
13
PDB header: transport proteinChain: A: PDB Molecule: sulfonate abc transporter, periplasmic sulfonate-bindingPDBTitle: crystal structure of putative sulfonate abc transporter, periplasmic2 sulfonate-binding protein ssua from methylobacillus flagellatus kt
104 c3l6gA_
not modelled
98.9
12
PDB header: glycine betaine-binding proteinChain: A: PDB Molecule: betaine abc transporter permease and substrate bindingPDBTitle: crystal structure of lactococcal opuac in its open conformation
105 c4oteA_
not modelled
98.7
9
PDB header: protein transportChain: A: PDB Molecule: lipoprotein;PDBTitle: crystal structure of a putative lipoprotein (cd630_1653) from2 clostridium difficile 630 at 2.20 a resolution
106 c3ix1B_
not modelled
98.7
11
PDB header: biosynthetic proteinChain: B: PDB Molecule: n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine bindingPDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
107 c4q5tA_
not modelled
98.7
11
PDB header: transport proteinChain: A: PDB Molecule: lipoprotein;PDBTitle: crystal structure of an atmb (putative membrane lipoprotein) from2 streptococcus mutans ua159 at 1.91 a resolution
108 c3ix1A_
not modelled
98.7
11
PDB header: biosynthetic proteinChain: A: PDB Molecule: n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine bindingPDBTitle: periplasmic n-formyl-4-amino-5-aminomethyl-2-methylpyrimidine binding2 protein from bacillus halodurans
109 d1xs5a_
not modelled
98.7
11
Fold: Periplasmic binding protein-like IISuperfamily: Periplasmic binding protein-like IIFamily: Phosphate binding protein-like
110 c3qslA_
not modelled
98.7
15
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: putative exported protein;PDBTitle: structure of cae31940 from bordetella bronchiseptica rb50
111 c3tqwA_
not modelled
98.6
12
PDB header: transport proteinChain: A: PDB Molecule: methionine-binding protein;PDBTitle: structure of a abc transporter, periplasmic substrate-binding protein2 from coxiella burnetii
112 c4ntlA_
not modelled
98.6
13
PDB header: transport proteinChain: A: PDB Molecule: lipoprotein, yaec family;PDBTitle: crystal structure of a lipoprotein, yaec family (ef3198) from2 enterococcus faecalis v583 at 1.80 a resolution
113 c3k2dA_
not modelled
98.6
13
PDB header: immune systemChain: A: PDB Molecule: abc-type metal ion transport system, periplasmic component;PDBTitle: crystal structure of immunogenic lipoprotein a from vibrio vulnificus
114 c6jf1A_
not modelled
98.6
12
PDB header: transport proteinChain: A: PDB Molecule: lipoprotein;PDBTitle: crystal structure of the substrate binding protein of a methionine2 transporter from streptococcus pneumoniae
115 c3hn0A_
not modelled
98.6
11
PDB header: transport proteinChain: A: PDB Molecule: nitrate transport protein;PDBTitle: crystal structure of an abc transporter (bdi_1369) from2 parabacteroides distasonis at 1.75 a resolution
116 c3tmgA_
not modelled
98.5
15
PDB header: transport proteinChain: A: PDB Molecule: glycine betaine, l-proline abc transporter,PDBTitle: crystal structure of glycine betaine, l-proline abc transporter,2 glycine/betaine/l-proline-binding protein (prox) from borrelia3 burgdorferi
117 c3up9A_
not modelled
98.5
13
PDB header: methionine-binding proteinChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of a putative lipoprotein (actodo_00931) from2 actinomyces odontolyticus atcc 17982 at 2.35 a resolution
118 c3un6A_
not modelled
98.4
9
PDB header: unknown functionChain: A: PDB Molecule: hypothetical protein saouhsc_00137;PDBTitle: 2.0 angstrom crystal structure of ligand binding component of abc-type2 import system from staphylococcus aureus with zinc bound
119 c4de8A_
not modelled
98.3
13
PDB header: membrane proteinChain: A: PDB Molecule: cps2a;PDBTitle: lytr-cps2a-psr family protein with bound octaprenyl monophosphate2 lipid
120 c4ef2A_
not modelled
98.3
11
PDB header: methionine-binding proteinChain: A: PDB Molecule: pheromone cob1/lipoprotein, yaec family;PDBTitle: crystal structure of a pheromone cob1 precursor/lipoprotein, yaec2 family (ef2496) from enterococcus faecalis v583 at 2.10 a resolution