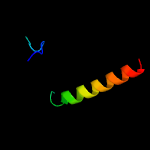
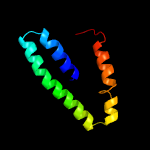
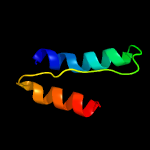
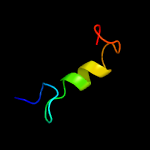
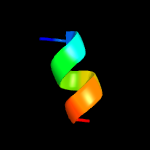
1 c5xuaB_
91.1
12
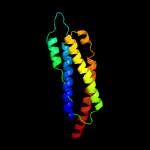
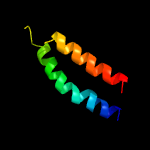
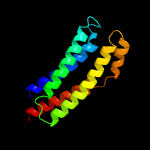
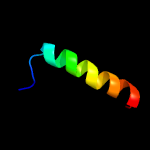
PDB header: signaling proteinChain: B: PDB Molecule: methyl-accepting chemotaxis sensory transducer;PDBTitle: the ligand-free dimer of chemoreceptor mcp2201 ligand binding domain
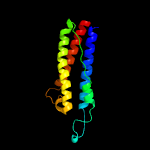
2 c4z9hA_
67.3
13
PDB header: protein bindingChain: A: PDB Molecule: methyl-accepting chemotaxis protein ii;PDBTitle: asp-tar from e. coli
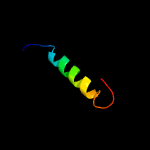
3 c5awwG_
60.5
39
PDB header: protein transport/immune systemChain: G: PDB Molecule: putative preprotein translocase, secg subunit;PDBTitle: precise resting state of thermus thermophilus secyeg
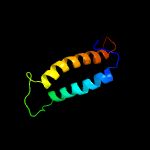
4 c2d4uA_
50.2
10
PDB header: signaling proteinChain: A: PDB Molecule: methyl-accepting chemotaxis protein i;PDBTitle: crystal structure of the ligand binding domain of the bacterial serine2 chemoreceptor tsr
5 c5nf8A_
43.7
16
PDB header: membrane proteinChain: A: PDB Molecule: respiratory supercomplex factor 1, mitochondrial;PDBTitle: solution structure of detergent-solubilized rcf1, a yeast2 mitochondrial inner membrane protein involved in respiratory complex3 iii/iv supercomplex formation
6 c3va9A_
38.3
11
PDB header: transferaseChain: A: PDB Molecule: sensor histidine kinase;PDBTitle: crystal structure of the rhodopseudomonas palustris histidine kinase2 hk9 sensor domain
7 c6f46A_
27.9
23
PDB header: apoptosisChain: A: PDB Molecule: bcl-2-like protein 1;PDBTitle: structure of the transmembrane helix of bclxl in phospholipid2 nanodiscs
8 c2mkvA_
26.3
22
PDB header: transport proteinChain: A: PDB Molecule: sodium/potassium-transporting atpase subunit gamma;PDBTitle: structure of the na,k-atpase regulatory protein fxyd2b in micelles
9 d2liga_
26.1
17
Fold: Four-helical up-and-down bundleSuperfamily: Aspartate receptor, ligand-binding domainFamily: Aspartate receptor, ligand-binding domain
10 c2kncA_
20.1
10
PDB header: cell adhesionChain: A: PDB Molecule: integrin alpha-iib;PDBTitle: platelet integrin alfaiib-beta3 transmembrane-cytoplasmic2 heterocomplex
11 c6hwhX_
19.9
15
PDB header: electron transportChain: X: PDB Molecule: cytochrome c oxidase polypeptide 4;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
12 c2lonA_
15.2
15
PDB header: membrane proteinChain: A: PDB Molecule: hig1 domain family member 1b;PDBTitle: backbone structure of human membrane protein higd1b
13 d2itba1
14.8
13
Fold: Ferritin-likeSuperfamily: Ferritin-likeFamily: MiaE-like
14 c6h9cD_
14.5
63
PDB header: virusChain: D: PDB Molecule: vp7;PDBTitle: cryo-em structure of archaeal extremophilic internal membrane-2 containing haloarcula californiae icosahedral virus 1 (hciv-1) at3 3.74 angstroms resolution.
15 c6f87C_
14.2
23
PDB header: transferaseChain: C: PDB Molecule: threonylcarbamoyl-amp synthase;PDBTitle: crystal structure of p. abyssi sua5 complexed with l-threonine and ppi
16 c1n7sA_
12.7
25
PDB header: transport proteinChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: high resolution structure of a truncated neuronal snare complex
17 c5ireD_
10.5
27
PDB header: virusChain: D: PDB Molecule: m protein;PDBTitle: the cryo-em structure of zika virus
18 c4xypA_
9.8
44
PDB header: viral proteinChain: A: PDB Molecule: fusion protein;PDBTitle: crystal structure of a piscine viral fusion protein
19 c6jimA_
9.7
20
PDB header: viral protein/rnaChain: A: PDB Molecule: helicase;PDBTitle: viral helicase protein
20 c5wsnD_
9.5
23
PDB header: virusChain: D: PDB Molecule: m protein;PDBTitle: structure of japanese encephalitis virus
21 c4qr8B_
not modelled
9.5
12
PDB header: hydrolaseChain: B: PDB Molecule: xaa-pro dipeptidase;PDBTitle: crystal structure of e coli pepq
22 c6adqP_
not modelled
9.0
28
PDB header: electron transportChain: P: PDB Molecule: prokaryotic respiratory supercomplex associate factor 1PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
23 c2mgxA_
not modelled
9.0
18
PDB header: unknown functionChain: A: PDB Molecule: steroid receptor rna activator 1;PDBTitle: nmr structure of sra1p c-terminal domain
24 c6an7D_
not modelled
8.7
7
PDB header: transport proteinChain: D: PDB Molecule: transport permease protein;PDBTitle: crystal structure of o-antigen polysaccharide abc-transporter
25 c6cc4A_
not modelled
8.4
19
PDB header: transport proteinChain: A: PDB Molecule: soluble cytochrome b562, lipid ii flippase murj chimera;PDBTitle: structure of murj from escherichia coli
26 c3j2pD_
not modelled
8.4
9
PDB header: viral proteinChain: D: PDB Molecule: small envelope protein m;PDBTitle: cryoem structure of dengue virus envelope protein heterotetramer
27 c3mzlH_
not modelled
7.9
19
PDB header: protein transportChain: H: PDB Molecule: protein transport protein sec31;PDBTitle: sec13/sec31 edge element, loop deletion mutant
28 c4wfaU_
not modelled
7.7
44
PDB header: ribosomeChain: U: PDB Molecule: 50s ribosomal protein l28;PDBTitle: the crystal structure of the large ribosomal subunit of staphylococcus2 aureus in complex with linezolid
29 c1qw1A_
not modelled
7.4
31
PDB header: gene regulationChain: A: PDB Molecule: diphtheria toxin repressor;PDBTitle: solution structure of the c-terminal domain of dtxr2 residues 110-226
30 c3mb2J_
not modelled
7.3
50
PDB header: isomeraseChain: J: PDB Molecule: 4-oxalocrotonate tautomerase family enzyme - beta subunit;PDBTitle: kinetic and structural characterization of a heterohexamer 4-2 oxalocrotonate tautomerase from chloroflexus aurantiacus j-10-fl:3 implications for functional and structural diversity in the4 tautomerase superfamily
31 c6hwhT_
not modelled
7.3
12
PDB header: electron transportChain: T: PDB Molecule: uncharacterized protein msmeg_4692/msmei_4575;PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
32 c1gl2A_
not modelled
7.0
29
PDB header: membrane proteinChain: A: PDB Molecule: endobrevin;PDBTitle: crystal structure of an endosomal snare core complex
33 c4cbfB_
not modelled
6.9
23
PDB header: virusChain: B: PDB Molecule: m protein;PDBTitle: near-atomic resolution cryo-em structure of dengue serotype 4 virus
34 c2pm7A_
not modelled
6.7
19
PDB header: protein transportChain: A: PDB Molecule: protein transport protein sec31;PDBTitle: crystal structure of yeast sec13/31 edge element of the copii2 vesicular coat, selenomethionine version
35 c2yruA_
not modelled
6.6
12
PDB header: apoptosisChain: A: PDB Molecule: steroid receptor rna activator 1;PDBTitle: solution structure of mouse steroid receptor rna activator2 1 (sra1) protein
36 c1sfcI_
not modelled
6.5
25
PDB header: transport proteinChain: I: PDB Molecule: protein (synaptobrevin 2);PDBTitle: neuronal synaptic fusion complex
37 c1p58E_
not modelled
6.5
9
PDB header: virusChain: E: PDB Molecule: envelope protein m;PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
38 c2lomA_
not modelled
6.5
6
PDB header: membrane proteinChain: A: PDB Molecule: hig1 domain family member 1a;PDBTitle: backbone structure of human membrane protein higd1a
39 c5ux1D_
not modelled
6.4
14
PDB header: lyaseChain: D: PDB Molecule: trna-(ms(2)io(6)a)-hydroxylase-like;PDBTitle: protein 43 with aldehyde deformylating oxygenase activity from2 synechococcus
40 d1hw1a2
not modelled
6.4
7
Fold: GntR ligand-binding domain-likeSuperfamily: GntR ligand-binding domain-likeFamily: GntR ligand-binding domain-like
41 c1urqA_
not modelled
6.2
18
PDB header: transport proteinChain: A: PDB Molecule: m-tomosyn isoform;PDBTitle: crystal structure of neuronal q-snares in complex with r-snare motif2 of tomosyn
42 c6an7C_
not modelled
6.2
7
PDB header: transport proteinChain: C: PDB Molecule: transport permease protein;PDBTitle: crystal structure of o-antigen polysaccharide abc-transporter
43 c3hd7A_
not modelled
6.1
25
PDB header: exocytosisChain: A: PDB Molecule: vesicle-associated membrane protein 2;PDBTitle: helical extension of the neuronal snare complex into the membrane,2 spacegroup c 1 2 1
44 d2hs5a2
not modelled
6.1
24
Fold: GntR ligand-binding domain-likeSuperfamily: GntR ligand-binding domain-likeFamily: GntR ligand-binding domain-like
45 c3dwlG_
not modelled
6.0
21
PDB header: structural proteinChain: G: PDB Molecule: actin-related protein 2/3 complex subunit 5;PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
46 d1jr1a4
not modelled
5.6
43
Fold: CBS-domain pairSuperfamily: CBS-domain pairFamily: CBS-domain pair
47 c5j12A_
not modelled
5.4
36
PDB header: signaling proteinChain: A: PDB Molecule: thymic stromal lymphopoietin;PDBTitle: structure of human tslp:tslpr in complex with mouse il-7ralpha
48 c3gw4B_
not modelled
5.3
15
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of uncharacterized protein from deinococcus2 radiodurans. northeast structural genomics consortium target drr162b.
49 c6gv9K_
not modelled
5.3
17
PDB header: protein fibrilChain: K: PDB Molecule: prepilin peptidase-dependent protein d;PDBTitle: structure of the type iv pilus from enterohemorrhagic escherichia coli2 (ehec)
50 c3b5nE_
not modelled
5.3
21
PDB header: membrane proteinChain: E: PDB Molecule: synaptobrevin homolog 1;PDBTitle: structure of the yeast plasma membrane snare complex
51 c2eqaA_
not modelled
5.2
18
PDB header: rna binding proteinChain: A: PDB Molecule: hypothetical protein st1526;PDBTitle: crystal structure of the hypothetical sua5 protein from2 sulfolobus tokodaii
52 d2axth1
not modelled
5.2
10
Fold: Single transmembrane helixSuperfamily: Photosystem II 10 kDa phosphoprotein PsbHFamily: PsbH-like
53 c2axtH_
not modelled
5.2
10
PDB header: electron transportChain: H: PDB Molecule: photosystem ii reaction center h protein;PDBTitle: crystal structure of photosystem ii from thermosynechococcus elongatus
54 d1ff3c_
not modelled
5.1
45
Fold: Ferredoxin-likeSuperfamily: Peptide methionine sulfoxide reductaseFamily: Peptide methionine sulfoxide reductase
55 c2vxgB_
not modelled
5.1
24
PDB header: gene regulationChain: B: PDB Molecule: cg6181-pa, isoform a;PDBTitle: crystal structure of the conserved c-terminal region of ge-2 1
56 c3csxA_
not modelled
5.0
35
PDB header: metal binding protein,unknown functionChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: structural characterization of a protein in the duf6832 family- crystal structure of cce_0567 from the3 cyanobacterium cyanothece 51142.