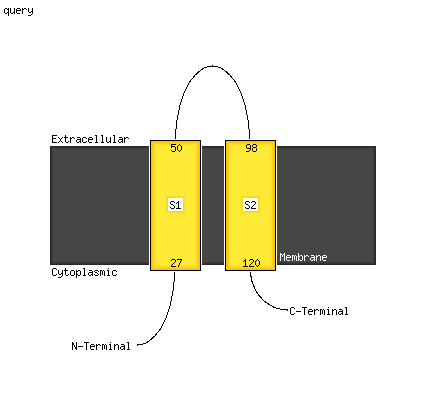
1 c5t42A_
58.0
48
PDB header: viral proteinChain: A: PDB Molecule: envelope glycoprotein;PDBTitle: structure of the ebola virus envelope protein mper/tm domain and its2 interaction with the fusion loop explains their fusion activity
2 c2lomA_
25.5
15
PDB header: membrane proteinChain: A: PDB Molecule: hig1 domain family member 1a;PDBTitle: backbone structure of human membrane protein higd1a
3 c5xnmj_
25.2
47
PDB header: membrane proteinChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: structure of unstacked c2s2m2-type psii-lhcii supercomplex from pisum2 sativum
4 c3jcuj_
19.4
42
PDB header: membrane proteinChain: J: PDB Molecule: photosystem ii reaction center protein j;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
5 c1p84E_
16.6
18
PDB header: oxidoreductaseChain: E: PDB Molecule: ubiquinol-cytochrome c reductase iron-sulfur subunit;PDBTitle: hdbt inhibited yeast cytochrome bc1 complex
6 c6dlnB_
16.5
23
PDB header: membrane proteinChain: B: PDB Molecule: transmembrane protein gp41;PDBTitle: oligomeric structure of the hiv gp41 mper-tmd in phospholipid bilayers
7 c5ydfA_
13.6
19
PDB header: transcriptionChain: A: PDB Molecule: parafibromin;PDBTitle: crystal structure of a disease-related gene, hcdc73(1-100)
8 c4gd3T_
12.9
18
PDB header: oxidoreductase/electron transportChain: T: PDB Molecule: hydrogenase-1 small chain;PDBTitle: structure of e. coli hydrogenase-1 in complex with cytochrome b
9 d1u2ca2
11.1
29
Fold: Dystroglycan, domain 2Superfamily: Dystroglycan, domain 2Family: Dystroglycan, domain 2
10 c5kk2E_
10.8
23
PDB header: membrane protein, transport protein, sigChain: E: PDB Molecule: voltage-dependent calcium channel gamma-2 subunit;PDBTitle: architecture of fully occupied glua2 ampa receptor - tarp complex2 elucidated by single particle cryo-electron microscopy
11 c2lonA_
10.6
17
PDB header: membrane proteinChain: A: PDB Molecule: hig1 domain family member 1b;PDBTitle: backbone structure of human membrane protein higd1b
12 c2k9yA_
10.1
20
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
13 c2k9yB_
10.1
20
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
14 d1r18a_
9.7
31
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Protein-L-isoaspartyl O-methyltransferase
15 d1fvia1
9.1
21
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: DNA ligase/mRNA capping enzyme postcatalytic domain
16 c2mi2A_
9.0
26
PDB header: transport proteinChain: A: PDB Molecule: sec-independent protein translocase protein tatb;PDBTitle: solution structure of the e. coli tatb protein in dpc micelles
17 c3jcuw_
8.4
40
PDB header: membrane proteinChain: W: PDB Molecule: photosystem ii reaction center w protein, chloroplastic;PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
18 c6q56C_
7.8
0
PDB header: rna binding proteinChain: C: PDB Molecule: trna (adenine(22)-n(1))-methyltransferase;PDBTitle: crystal structure of the b. subtilis m1a22 trna methyltransferase trmk
19 c3jbrE_
7.7
18
PDB header: membrane proteinChain: E: PDB Molecule: voltage-dependent calcium channel gamma-1 subunit;PDBTitle: cryo-em structure of the rabbit voltage-gated calcium channel cav1.12 complex at 4.2 angstrom
20 c6ithA_
7.7
18
PDB header: membrane proteinChain: A: PDB Molecule: syndecan-2;PDBTitle: structure of the transmembrane domain of syndecan 2 in micelles
21 c6ckgA_
not modelled
7.6
12
PDB header: transferaseChain: A: PDB Molecule: d-glycerate 3-kinase;PDBTitle: d-glycerate 3-kinase from cryptococcus neoformans var. grubii serotype2 a (h99 / atcc 208821 / cbs 10515 / fgsc 9487)
22 d2gpia1
not modelled
7.5
29
Fold: Shew3726-likeSuperfamily: Shew3726-likeFamily: Shew3726-like
23 c4alyB_
not modelled
7.4
20
PDB header: membrane proteinChain: B: PDB Molecule: p35 antigen;PDBTitle: borrelia burgdorferi outer surface lipoprotein bba64
24 d1vbfa_
not modelled
7.2
16
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Protein-L-isoaspartyl O-methyltransferase
25 c2d7uA_
not modelled
7.1
6
PDB header: ligaseChain: A: PDB Molecule: adenylosuccinate synthetase;PDBTitle: crystal structure of hypothetical adenylosuccinate synthetase, ph04382 from pyrococcus horikoshii ot3
26 c2n9oA_
not modelled
7.0
44
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase rnf126;PDBTitle: solution structure of rnf126 n-terminal zinc finger domain
27 c6e8wC_
not modelled
6.9
23
PDB header: viral proteinChain: C: PDB Molecule: envelope glycoprotein gp160;PDBTitle: mper-tm domain of hiv-1 envelope glycoprotein (env)
28 c5xlxD_
not modelled
6.8
38
PDB header: transferaseChain: D: PDB Molecule: chemotaxis protein methyltransferase 1;PDBTitle: crystal structure of the c-terminal domain of cher1 containing sah
29 c5ftwA_
not modelled
6.8
25
PDB header: transferaseChain: A: PDB Molecule: chemotaxis protein methyltransferase;PDBTitle: crystal structure of glutamate o-methyltransferase in2 complex with s- adenosyl-l-homocysteine (sah) from3 bacillus subtilis
30 c6fmlJ_
not modelled
6.7
11
PDB header: dna binding proteinChain: J: PDB Molecule: actin related protein 5;PDBTitle: cryoem structure ino80core nucleosome complex
31 c5nf8A_
not modelled
6.6
21
PDB header: membrane proteinChain: A: PDB Molecule: respiratory supercomplex factor 1, mitochondrial;PDBTitle: solution structure of detergent-solubilized rcf1, a yeast2 mitochondrial inner membrane protein involved in respiratory complex3 iii/iv supercomplex formation
32 d1af7a2
not modelled
6.2
13
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Chemotaxis receptor methyltransferase CheR, C-terminal domain
33 c5fcdA_
not modelled
6.1
33
PDB header: transferaseChain: A: PDB Molecule: mccd;PDBTitle: crystal structure of mccd protein
34 c3gnlB_
not modelled
6.0
0
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein, duf633, lmof2365_1472;PDBTitle: structure of uncharacterized protein (lmof2365_1472) from listeria2 monocytogenes serotype 4b
35 d1jsxa_
not modelled
5.8
33
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Glucose-inhibited division protein B (GidB)
36 c4p7cB_
not modelled
5.6
26
PDB header: transferaseChain: B: PDB Molecule: trna (mo5u34)-methyltransferase;PDBTitle: crystal structure of putative methyltransferase from pseudomonas2 syringae pv. tomato
37 c2pbfA_
not modelled
5.5
38
PDB header: transferaseChain: A: PDB Molecule: protein-l-isoaspartate o-methyltransferase beta-aspartatePDBTitle: crystal structure of a putative protein-l-isoaspartate o-2 methyltransferase beta-aspartate methyltransferase (pcmt) from3 plasmodium falciparum in complex with s-adenosyl-l-homocysteine
38 d1jg1a_
not modelled
5.4
45
Fold: S-adenosyl-L-methionine-dependent methyltransferasesSuperfamily: S-adenosyl-L-methionine-dependent methyltransferasesFamily: Protein-L-isoaspartyl O-methyltransferase
39 c4d05A_
not modelled
5.4
21
PDB header: ligaseChain: A: PDB Molecule: atp-dependent dna ligase;PDBTitle: structure and activity of a minimal-type atp-dependent dna ligase from2 a psychrotolerant bacterium
40 d1knza_
not modelled
5.2
41
Fold: NSP3 homodimerSuperfamily: NSP3 homodimerFamily: NSP3 homodimer
41 c5w7kA_
not modelled
5.2
21
PDB header: transferaseChain: A: PDB Molecule: oxag;PDBTitle: crystal structure of oxag