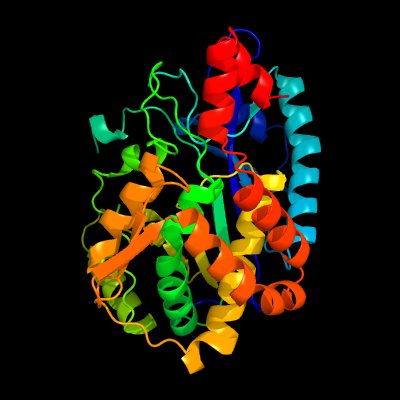
| 1 | d1r8ga_
|
|
 |
100.0 |
28 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:Glutamate-cysteine ligase family 2 (GCS2) |
|
|
|
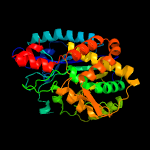
| 2 | d1tt4a_
|
|
 |
100.0 |
27 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:Glutamate-cysteine ligase family 2 (GCS2) |
|
|
|
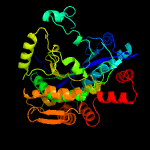
| 3 | c1tt4B_
|
|
 |
100.0 |
27 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:putative cytoplasmic protein;
PDBTitle: structure of np459575, a predicted glutathione synthase from2 salmonella typhimurium
|
|
|
|
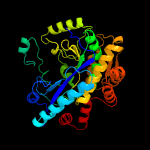
| 4 | c2gwcE_
|
|
 |
100.0 |
21 |
PDB header:ligase
Chain: E: PDB Molecule:glutamate cysteine ligase;
PDBTitle: crystal structure of plant glutamate cysteine ligase in complex with a2 transition state analogue
|
|
|
|
| 5 | c3nztA_
|
|
 |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:glutamate--cysteine ligase;
PDBTitle: 2.0 angstrom crystal structure of glutamate--cysteine ligase (gsha)2 ftom francisella tularensis in complex with amp
|
|
|
|
| 6 | d2d32a1
|
|
 |
100.0 |
15 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:Glutamate-cysteine ligase |
|
|
|
| 7 | c3ln7A_
|
|
 |
100.0 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:glutathione biosynthesis bifunctional protein gshab;
PDBTitle: crystal structure of a bifunctional glutathione synthetase from2 pasteurella multocida
|
|
|
|
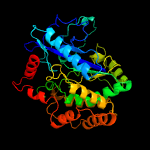
| 8 | c3ln6A_
|
|
 |
100.0 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:glutathione biosynthesis bifunctional protein gshab;
PDBTitle: crystal structure of a bifunctional glutathione synthetase from2 streptococcus agalactiae
|
|
|
|
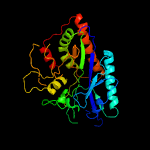
| 9 | c3lvwA_
|
|
 |
99.8 |
21 |
PDB header:ligase
Chain: A: PDB Molecule:glutamate--cysteine ligase;
PDBTitle: glutathione-inhibited scgcl
|
|
|
|
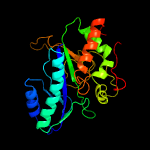
| 10 | c4hppA_
|
|
 |
99.2 |
18 |
PDB header:ligase
Chain: A: PDB Molecule:probable glutamine synthetase;
PDBTitle: crystal structure of novel glutamine synthase homolog
|
|
|
|
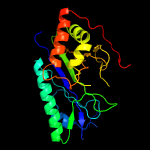
| 11 | c5zlpH_
|
|
 |
99.1 |
22 |
PDB header:ligase
Chain: H: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of glutamine synthetase from helicobacter pylori
|
|
|
|
| 12 | d2bvca2
|
|
 |
99.1 |
20 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:Glutamine synthetase catalytic domain |
|
|
|
| 13 | d1f52a2
|
|
 |
99.0 |
19 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:Glutamine synthetase catalytic domain |
|
|
|
| 14 | c1fpyE_
|
|
 |
99.0 |
19 |
PDB header:ligase
Chain: E: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of glutamine synthetase from salmonella2 typhimurium with inhibitor phosphinothricin
|
|
|
|
| 15 | c3ng0A_
|
|
 |
99.0 |
20 |
PDB header:ligase
Chain: A: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of glutamine synthetase from synechocystis sp. pcc2 6803
|
|
|
|
| 16 | c4s17E_
|
|
 |
99.0 |
22 |
PDB header:ligase
Chain: E: PDB Molecule:glutamine synthetase;
PDBTitle: the crystal structure of glutamine synthetase from bifidobacterium2 adolescentis atcc 15703
|
|
|
|
| 17 | c1htoB_
|
|
 |
98.9 |
20 |
PDB header:ligase
Chain: B: PDB Molecule:glutamine synthetase;
PDBTitle: crystallographic structure of a relaxed glutamine synthetase from2 mycobacterium tuberculosis
|
|
|
|
| 18 | c5dm3A_
|
|
 |
98.9 |
15 |
PDB header:ligase
Chain: A: PDB Molecule:l-glutamine synthetase;
PDBTitle: crystal structure of glutamine synthetase from chromohalobacter2 salexigens dsm 3043(csal_0679, target efi-550015) with bound adp
|
|
|
|
| 19 | c3qajL_
|
|
 |
98.9 |
17 |
PDB header:ligase
Chain: L: PDB Molecule:glutamine synthetase;
PDBTitle: x-ray crystal structure of glutamine synthetase from bacillus subtilis2 cocrystallized with atp
|
|
|
|
| 20 | c3o6xC_
|
|
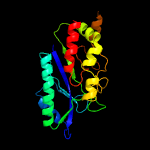 |
98.7 |
20 |
PDB header:ligase
Chain: C: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of the type iii glutamine synthetase from2 bacteroides fragilis
|
|
|
|
| 21 | c4is4G_ |
|
not modelled |
98.5 |
17 |
PDB header:ligase
Chain: G: PDB Molecule:glutamine synthetase;
PDBTitle: the glutamine synthetase from the dicotyledonous plant m. truncatula2 is a decamer
|
|
|
| 22 | c4baxH_ |
|
not modelled |
98.4 |
15 |
PDB header:ligase
Chain: H: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of glutamine synthetase from streptomyces2 coelicolor
|
|
|
| 23 | c2j9iL_ |
|
not modelled |
98.4 |
18 |
PDB header:ligase
Chain: L: PDB Molecule:glutamate-ammonia ligase domain-containing protein 1;
PDBTitle: lengsin is a survivor of an ancient family of class i glutamine2 synthetases in eukaryotes that has undergone evolutionary re-3 engineering for a tissue-specific role in the vertebrate eye lens.
|
|
|
| 24 | c2qc8J_ |
|
not modelled |
98.3 |
19 |
PDB header:ligase
Chain: J: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of human glutamine synthetase in complex with adp2 and methionine sulfoximine phosphate
|
|
|
| 25 | c2d3aJ_ |
|
not modelled |
98.2 |
18 |
PDB header:ligase
Chain: J: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of the maize glutamine synthetase2 complexed with adp and methionine sulfoximine phosphate
|
|
|
| 26 | c3fkyD_ |
|
not modelled |
97.7 |
22 |
PDB header:ligase
Chain: D: PDB Molecule:glutamine synthetase;
PDBTitle: crystal structure of the glutamine synthetase gln1deltan182 from the yeast saccharomyces cerevisiae
|
|
|
| 27 | c4bjrA_ |
|
not modelled |
93.3 |
13 |
PDB header:ligase
Chain: A: PDB Molecule:pup--protein ligase, prokaryotic ubiquitin-like protein
PDBTitle: crystal structure of the complex between prokaryotic2 ubiquitin-like protein pup and its ligase pafa
|
|
|
| 28 | c4b0tB_ |
|
not modelled |
68.2 |
11 |
PDB header:ligase
Chain: B: PDB Molecule:pup--protein ligase;
PDBTitle: structure of the pup ligase pafa of the prokaryotic2 ubiquitin-like modification pathway in complex with adp
|
|
|
| 29 | c4b0sA_ |
|
not modelled |
45.5 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:deamidase-depupylase dop;
PDBTitle: structure of the deamidase-depupylase dop of the prokaryotic2 ubiquitin-like modification pathway in complex with atp
|
|
|
| 30 | c2ebbA_ |
|
not modelled |
44.6 |
13 |
PDB header:lyase
Chain: A: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine2 dehydratase (pterin carbinolamine dehydratase) from3 geobacillus kaustophilus hta426
|
|
|
| 31 | c2v6uB_ |
|
not modelled |
30.2 |
12 |
PDB header:lyase
Chain: B: PDB Molecule:pterin-4a-carbinolamine dehydratase;
PDBTitle: high resolution crystal structure of pterin-4a-2 carbinolamine dehydratase from toxoplasma gondii
|
|
|
| 32 | d1ru0a_ |
|
not modelled |
29.9 |
12 |
Fold:DCoH-like
Superfamily:PCD-like
Family:PCD-like |
|
|
| 33 | d1dcpa_ |
|
not modelled |
23.5 |
9 |
Fold:DCoH-like
Superfamily:PCD-like
Family:PCD-like |
|
|
| 34 | c3jstA_ |
|
not modelled |
15.0 |
12 |
PDB header:lyase
Chain: A: PDB Molecule:putative pterin-4-alpha-carbinolamine dehydratase;
PDBTitle: crystal structure of transcriptional coactivator/pterin dehydratase2 from brucella melitensis
|
|
|
| 35 | d2pw6a1 |
|
not modelled |
13.4 |
19 |
Fold:Phosphorylase/hydrolase-like
Superfamily:LigB-like
Family:LigB-like |
|
|
| 36 | c3oqhB_ |
|
not modelled |
13.4 |
15 |
PDB header:ligase
Chain: B: PDB Molecule:putative uncharacterized protein yvmc;
PDBTitle: crystal structure of b. licheniformis cdps yvmc-blic
|
|
|
| 37 | d3ci0k1 |
|
not modelled |
12.5 |
14 |
Fold:SAM domain-like
Superfamily:GspK insert domain-like
Family:GspK insert domain-like |
|
|
| 38 | c2p7pB_ |
|
not modelled |
12.4 |
16 |
PDB header:metal binding protein, hydrolase
Chain: B: PDB Molecule:glyoxalase family protein;
PDBTitle: crystal structure of genomically encoded fosfomycin resistance2 protein, fosx, from listeria monocytogenes complexed with mn(ii) and3 sulfate ion
|
|
|
| 39 | c5vb0E_ |
|
not modelled |
11.3 |
9 |
PDB header:transferase
Chain: E: PDB Molecule:fosfomycin resistance protein fosa3;
PDBTitle: crystal structure of fosfomycin resistance protein fosa3
|
|
|
| 40 | c2cp8A_ |
|
not modelled |
10.5 |
24 |
PDB header:protein binding
Chain: A: PDB Molecule:next to brca1 gene 1 protein;
PDBTitle: solution structure of the rsgi ruh-046, a uba domain from2 human next to brca1 gene 1 protein (kiaa0049 protein)3 r923h variant
|
|
|
| 41 | d1nkia_ |
|
not modelled |
9.7 |
13 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Antibiotic resistance proteins |
|
|
| 42 | d2eg6a1 |
|
not modelled |
9.6 |
21 |
Fold:TIM beta/alpha-barrel
Superfamily:Metallo-dependent hydrolases
Family:Dihydroorotase |
|
|
| 43 | d2d6fc3 |
|
not modelled |
9.4 |
15 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:GatB/GatE catalytic domain-like |
|
|
| 44 | d1usma_ |
|
not modelled |
9.0 |
19 |
Fold:DCoH-like
Superfamily:PCD-like
Family:PCD-like |
|
|
| 45 | c2krkA_ |
|
not modelled |
8.6 |
16 |
PDB header:protein binding
Chain: A: PDB Molecule:26s protease regulatory subunit 8;
PDBTitle: solution nmr structure of 26s protease regulatory subunit 82 from h.sapiens, northeast structural genomics consortium3 target target hr3102a
|
|
|
| 46 | c1g5jB_ |
|
not modelled |
8.3 |
67 |
PDB header:apoptosis
Chain: B: PDB Molecule:bad protein;
PDBTitle: complex of bcl-xl with peptide from bad
|
|
|
| 47 | d1r9ca_ |
|
not modelled |
7.9 |
20 |
Fold:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Superfamily:Glyoxalase/Bleomycin resistance protein/Dihydroxybiphenyl dioxygenase
Family:Antibiotic resistance proteins |
|
|
| 48 | c3rj1G_ |
|
not modelled |
7.4 |
38 |
PDB header:transcription
Chain: G: PDB Molecule:mediator of rna polymerase ii transcription subunit 6;
PDBTitle: architecture of the mediator head module
|
|
|
| 49 | d2qkwa1 |
|
not modelled |
7.1 |
13 |
Fold:immunoglobulin/albumin-binding domain-like
Superfamily:Avirulence protein AvrPto
Family:Avirulence protein AvrPto |
|
|
| 50 | c2qkwA_ |
|
not modelled |
7.1 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:avirulence protein;
PDBTitle: structural basis for activation of plant immunity by2 bacterial effector protein avrpto
|
|
|
| 51 | c4gwpG_ |
|
not modelled |
6.9 |
33 |
PDB header:transcription
Chain: G: PDB Molecule:mediator of rna polymerase ii transcription subunit 6;
PDBTitle: structure of the mediator head module from s. cerevisiae
|
|
|
| 52 | d1j5ya2 |
|
not modelled |
6.8 |
18 |
Fold:HPr-like
Superfamily:Putative transcriptional regulator TM1602, C-terminal domain
Family:Putative transcriptional regulator TM1602, C-terminal domain |
|
|
| 53 | c3zw5A_ |
|
not modelled |
6.7 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:glyoxalase domain-containing protein 5;
PDBTitle: crystal structure of the human glyoxalase domain-containing protein 5
|
|
|
| 54 | c6bu2A_ |
|
not modelled |
6.6 |
16 |
PDB header:isomerase
Chain: A: PDB Molecule:glyoxalase;
PDBTitle: crystal structure of methylmalonyl-coa epimerase from mycobacterium2 tuberculosis
|
|
|
| 55 | c5oevB_ |
|
not modelled |
6.6 |
10 |
PDB header:transferase
Chain: B: PDB Molecule:glutathione synthetase-like effector 22 (gpa-gss22-apo);
PDBTitle: the structure of a glutathione synthetase like-effector (gss22) from2 globodera pallida in apoform.
|
|
|
| 56 | c4eacC_ |
|
not modelled |
6.6 |
7 |
PDB header:lyase
Chain: C: PDB Molecule:mannonate dehydratase;
PDBTitle: crystal structure of mannonate dehydratase from escherichia coli2 strain k12
|
|
|
| 57 | d2f2ab2 |
|
not modelled |
6.5 |
26 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:GatB/GatE catalytic domain-like |
|
|
| 58 | c4n0iB_ |
|
not modelled |
6.4 |
11 |
PDB header:ligase
Chain: B: PDB Molecule:glutamyl-trna(gln) amidotransferase subunit b,
PDBTitle: crystal structure of s. cerevisiae mitochondrial gatfab in complex2 with glutamine
|
|
|
| 59 | c4hc5A_ |
|
not modelled |
6.3 |
12 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glyoxalase/bleomycin resistance protein/dioxygenase;
PDBTitle: crystal structure of member of glyoxalase/bleomycin resistance2 protein/dioxygenase superfamily from sphaerobacter thermophilus dsm3 20745
|
|
|
| 60 | c6avjB_ |
|
not modelled |
6.1 |
22 |
PDB header:metal binding protein
Chain: B: PDB Molecule:cdgsh iron-sulfur domain-containing protein 3,
PDBTitle: crystal structure of human mitochondrial inner neet protein (mint)2 /cisd3
|
|
|
| 61 | d1ylea1 |
|
not modelled |
6.1 |
43 |
Fold:Acyl-CoA N-acyltransferases (Nat)
Superfamily:Acyl-CoA N-acyltransferases (Nat)
Family:AstA-like |
|
|
| 62 | c2q25C_ |
|
not modelled |
5.9 |
9 |
PDB header:virus,hydrolase/rna
Chain: C: PDB Molecule:protein alpha;
PDBTitle: flock house virus coat protein d75n mutant
|
|
|
| 63 | c5zet2_ |
|
not modelled |
5.9 |
33 |
PDB header:ribosome
Chain: 2: PDB Molecule:50s ribosomal protein l31;
PDBTitle: m. smegmatis p/p state 50s ribosomal subunit
|
|
|
| 64 | c5zcrB_ |
|
not modelled |
5.7 |
11 |
PDB header:hydrolase
Chain: B: PDB Molecule:maltooligosyl trehalose synthase;
PDBTitle: dsm5389 glycosyltrehalose synthase
|
|
|
| 65 | c3tbmA_ |
|
not modelled |
5.7 |
33 |
PDB header:metal binding protein
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a type 4 cdgsh iron-sulfur protein.
|
|
|
| 66 | d1zq1c3 |
|
not modelled |
5.7 |
15 |
Fold:Glutamine synthetase/guanido kinase
Superfamily:Glutamine synthetase/guanido kinase
Family:GatB/GatE catalytic domain-like |
|
|
| 67 | c2bzwB_ |
|
not modelled |
5.6 |
67 |
PDB header:transcription
Chain: B: PDB Molecule:bcl2-antagonist of cell death;
PDBTitle: the crystal structure of bcl-xl in complex with full-length bad
|
|
|
| 68 | d2dnaa1 |
|
not modelled |
5.6 |
20 |
Fold:RuvA C-terminal domain-like
Superfamily:UBA-like
Family:UBA domain |
|
|
| 69 | c3jzeC_ |
|
not modelled |
5.4 |
21 |
PDB header:hydrolase
Chain: C: PDB Molecule:dihydroorotase;
PDBTitle: 1.8 angstrom resolution crystal structure of dihydroorotase (pyrc)2 from salmonella enterica subsp. enterica serovar typhimurium str. lt2
|
|
|
| 70 | c3bdkB_ |
|
not modelled |
5.4 |
18 |
PDB header:lyase
Chain: B: PDB Molecule:d-mannonate dehydratase;
PDBTitle: crystal structure of streptococcus suis mannonate2 dehydratase complexed with substrate analogue
|
|
|
| 71 | d1dpsa_ |
|
not modelled |
5.1 |
5 |
Fold:Ferritin-like
Superfamily:Ferritin-like
Family:Ferritin |
|
|