| 1 |
|
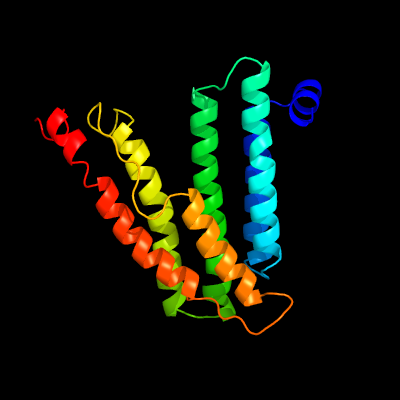
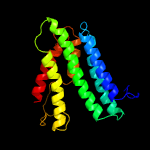
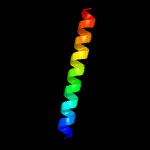
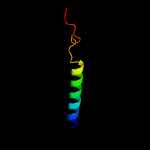
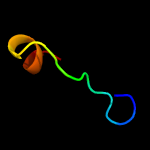
PDB 6h59 chain B
Region: 5 - 198
Aligned: 188
Modelled: 194
Confidence: 99.9%
Identity: 15%
PDB header:transferase
Chain: B: PDB Molecule:cdp-diacylglycerol--inositol 3-phosphatidyltransferase;
PDBTitle: crystal structure of mycobacterium tuberculosis phosphatidylinositol2 phosphate synthase (pgsa1) with cdp-dag bound
Phyre2
| 2 |
|
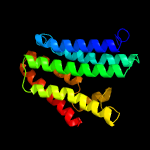
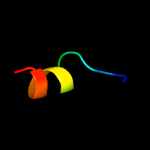
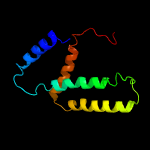
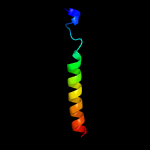
PDB 5d92 chain B
Region: 3 - 185
Aligned: 178
Modelled: 183
Confidence: 99.9%
Identity: 17%
PDB header:membrane protein
Chain: B: PDB Molecule:af2299 protein,phosphatidylinositol synthase;
PDBTitle: structure of a phosphatidylinositolphosphate (pip) synthase from2 renibacterium salmoninarum
Phyre2
| 3 |
|
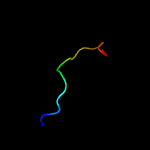
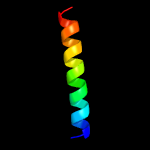
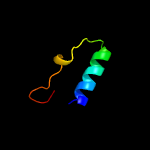
PDB 4o6m chain A
Region: 8 - 185
Aligned: 167
Modelled: 178
Confidence: 99.9%
Identity: 17%
PDB header:transferase
Chain: A: PDB Molecule:af2299, a cdp-alcohol phosphotransferase;
PDBTitle: structure of af2299, a cdp-alcohol phosphotransferase (cmp-bound)
Phyre2
| 4 |
|
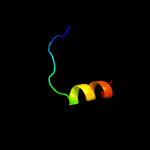
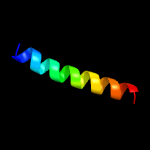
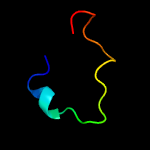
PDB 4mnd chain A
Region: 9 - 182
Aligned: 165
Modelled: 174
Confidence: 99.9%
Identity: 15%
PDB header:transferase
Chain: A: PDB Molecule:ctp l-myo-inositol-1-phosphate cytidylyltransferase/cdp-l-
PDBTitle: crystal structure of archaeoglobus fulgidus ipct-dipps bifunctional2 membrane protein
Phyre2
| 5 |
|
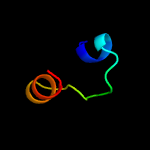
PDB 2cx6 chain A domain 1
Region: 59 - 82
Aligned: 24
Modelled: 23
Confidence: 32.7%
Identity: 17%
Fold: Barstar-like
Superfamily: Barstar-related
Family: Barstar-related
Phyre2
| 6 |
|
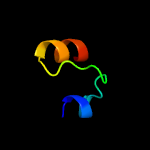
PDB 1ay7 chain B
Region: 59 - 82
Aligned: 24
Modelled: 24
Confidence: 28.4%
Identity: 21%
Fold: Barstar-like
Superfamily: Barstar-related
Family: Barstar-related
Phyre2
| 7 |
|
PDB 4i0x chain J
Region: 53 - 82
Aligned: 30
Modelled: 30
Confidence: 21.2%
Identity: 17%
PDB header:structural genomics, unknown function
Chain: J: PDB Molecule:esat-6-like protein mab_3113;
PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
Phyre2
| 8 |
|
PDB 2le7 chain A
Region: 59 - 72
Aligned: 14
Modelled: 14
Confidence: 11.0%
Identity: 64%
PDB header:transport protein
Chain: A: PDB Molecule:potassium voltage-gated channel subfamily h member 2;
PDBTitle: solution nmr structure of the s4s5 linker of herg potassium channel
Phyre2
| 9 |
|
PDB 2kt5 chain B
Region: 268 - 282
Aligned: 15
Modelled: 15
Confidence: 10.8%
Identity: 40%
PDB header:rna binding protein / viral protein
Chain: B: PDB Molecule:icp27;
PDBTitle: rrm domain of mrna export adaptor ref2-i bound to hsv-1 icp27 peptide
Phyre2
| 10 |
|
PDB 6mjb chain C
Region: 62 - 80
Aligned: 19
Modelled: 19
Confidence: 10.0%
Identity: 26%
PDB header:cell cycle
Chain: C: PDB Molecule:kinetochore-associated protein dsn1;
PDBTitle: structure of candida glabrata csm1:dsn1(14-72) complex
Phyre2
| 11 |
|
PDB 1w8x chain P
Region: 222 - 255
Aligned: 34
Modelled: 34
Confidence: 9.5%
Identity: 15%
PDB header:virus
Chain: P: PDB Molecule:protein p16;
PDBTitle: structural analysis of prd1
Phyre2
| 12 |
|
PDB 3lrc chain C
Region: 139 - 247
Aligned: 109
Modelled: 109
Confidence: 8.2%
Identity: 15%
PDB header:transport protein
Chain: C: PDB Molecule:arginine/agmatine antiporter;
PDBTitle: structure of e. coli adic (p1)
Phyre2
| 13 |
|
PDB 1gl2 chain D
Region: 53 - 76
Aligned: 24
Modelled: 24
Confidence: 6.4%
Identity: 29%
PDB header:membrane protein
Chain: D: PDB Molecule:syntaxin 8;
PDBTitle: crystal structure of an endosomal snare core complex
Phyre2
| 14 |
|
PDB 3b5n chain L
Region: 53 - 76
Aligned: 24
Modelled: 24
Confidence: 6.0%
Identity: 29%
PDB header:membrane protein
Chain: L: PDB Molecule:protein transport protein sec9;
PDBTitle: structure of the yeast plasma membrane snare complex
Phyre2
| 15 |
|
PDB 2ov2 chain O
Region: 131 - 148
Aligned: 18
Modelled: 18
Confidence: 5.7%
Identity: 17%
PDB header:protein binding/transferase
Chain: O: PDB Molecule:serine/threonine-protein kinase pak 4;
PDBTitle: the crystal structure of the human rac3 in complex with the crib2 domain of human p21-activated kinase 4 (pak4)
Phyre2
| 16 |
|
PDB 6h2e chain P
Region: 53 - 82
Aligned: 30
Modelled: 30
Confidence: 5.6%
Identity: 23%
PDB header:toxin
Chain: P: PDB Molecule:ahlc;
PDBTitle: structure of the soluble ahlc of the tripartite alpha-pore forming2 toxin, ahl, from aeromonas hydrophila.
Phyre2
| 17 |
|
PDB 1y0k chain A domain 1
Region: 237 - 268
Aligned: 32
Modelled: 32
Confidence: 5.1%
Identity: 13%
Fold: Restriction endonuclease-like
Superfamily: Restriction endonuclease-like
Family: PA4535-like
Phyre2
| 18 |
|
PDB 2amw chain A
Region: 55 - 78
Aligned: 24
Modelled: 24
Confidence: 5.1%
Identity: 29%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:hypothetical protein ne2163;
PDBTitle: solution nmr structure of protein ne2163 from nitrosomonas europaea.2 northeast structural genomics consortium target net1.
Phyre2