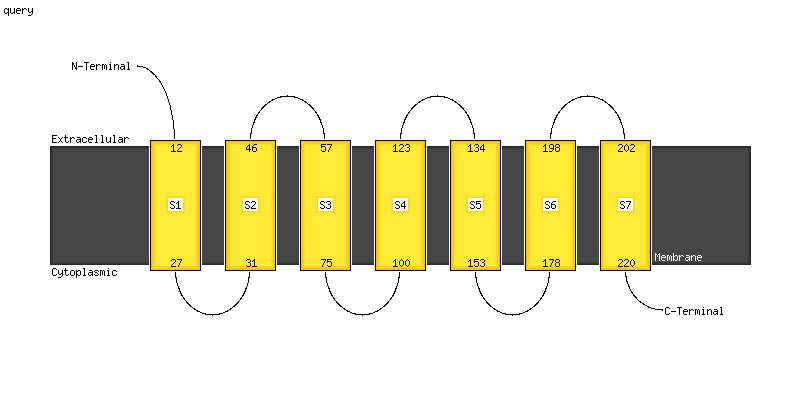
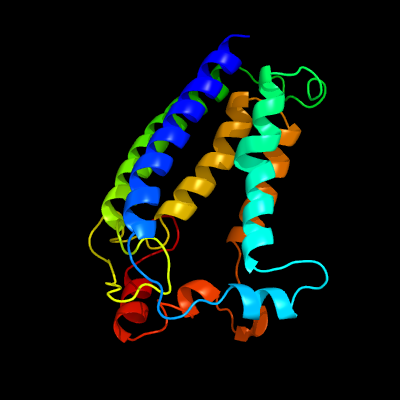
| 1 | c4a2nB_
|
|
 |
99.9 |
12 |
PDB header:transferase
Chain: B: PDB Molecule:isoprenylcysteine carboxyl methyltransferase;
PDBTitle: crystal structure of ma-icmt
|
|
|
|
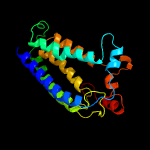
| 2 | c4quvB_
|
|
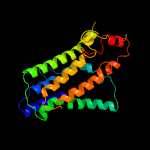 |
99.9 |
17 |
PDB header:oxidoreductase, membrane protein
Chain: B: PDB Molecule:delta(14)-sterol reductase;
PDBTitle: structure of an integral membrane delta(14)-sterol reductase
|
|
|
|
| 3 | c5v7pA_
|
|
 |
99.9 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:protein-s-isoprenylcysteine o-methyltransferase;
PDBTitle: atomic structure of the eukaryotic intramembrane ras methyltransferase2 icmt (isoprenylcysteine carboxyl methyltransferase), in complex with3 a monobody
|
|
|
|
| 4 | c4y9iA_
|
|
 |
72.1 |
25 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:mycobacterium tuberculosis paralogous family 11;
PDBTitle: structure of f420-h2 dependent reductase (fdr-a) msmeg_2027
|
|
|
|
| 5 | c3r5yC_
|
|
 |
68.0 |
32 |
PDB header:unknown function
Chain: C: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of a deazaflavin-dependent nitroreductase from nocardia2 farcinica, with co-factor f420
|
|
|
|
| 6 | c3r5wO_
|
|
 |
60.5 |
21 |
PDB header:oxidoreductase
Chain: O: PDB Molecule:deazaflavin-dependent nitroreductase;
PDBTitle: structure of ddn, the deazaflavin-dependent nitroreductase from2 mycobacterium tuberculosis involved in bioreductive activation of pa-3 824, with co-factor f420
|
|
|
|
| 7 | c3h96B_
|
|
 |
56.2 |
32 |
PDB header:flavoprotein
Chain: B: PDB Molecule:f420-h2 dependent reductase a;
PDBTitle: msmeg_3358 f420 reductase
|
|
|
|
| 8 | c3r5zB_
|
|
 |
54.6 |
29 |
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: structure of a deazaflavin-dependent reductase from nocardia2 farcinica, with co-factor f420
|
|
|
|
| 9 | c6hwhX_
|
|
 |
46.7 |
23 |
PDB header:electron transport
Chain: X: PDB Molecule:cytochrome c oxidase polypeptide 4;
PDBTitle: structure of a functional obligate respiratory supercomplex from2 mycobacterium smegmatis
|
|
|
|
| 10 | c4p63A_
|
|
 |
35.8 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:probable deoxyhypusine synthase;
PDBTitle: crystal structure of deoxyhypusine synthase from pyrococcus horikoshii
|
|
|
|
| 11 | d1e7la1
|
|
 |
32.9 |
56 |
Fold:LEM/SAP HeH motif
Superfamily:Recombination endonuclease VII, C-terminal and dimerization domains
Family:Recombination endonuclease VII, C-terminal and dimerization domains |
|
|
|
| 12 | c6dftJ_
|
|
 |
26.6 |
18 |
PDB header:transferase
Chain: J: PDB Molecule:deoxyhypusine synthase regulatory subunit;
PDBTitle: trypanosoma brucei deoxyhypusine synthase
|
|
|
|
| 13 | c5ht7A_
|
|
 |
24.1 |
27 |
PDB header:metal binding protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: crystal structure of a transition-metal-ion-binding betagamma-2 crystallin from methanosaeta thermophila
|
|
|
|
| 14 | c4hxhC_
|
|
 |
23.5 |
17 |
PDB header:rna/rna binding protein/hydrolase
Chain: C: PDB Molecule:histone rna hairpin-binding protein;
PDBTitle: structure of mrna stem-loop, human stem-loop binding protein and2 3'hexo ternary complex
|
|
|
|
| 15 | c5e8jC_
|
|
 |
22.1 |
19 |
PDB header:translation
Chain: C: PDB Molecule:rnmt-activating mini protein;
PDBTitle: crystal structure of mrna cap guanine-n7 methyltransferase in complex2 with ram
|
|
|
|
| 16 | c4q2eA_
|
|
 |
16.0 |
11 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidate cytidylyltransferase;
PDBTitle: crystal structure of an intramembrane cdp-dag synthetase central for2 phospholipid biosynthesis (s200c/s258c, active mutant)
|
|
|
|
| 17 | c6cebP_
|
|
 |
14.6 |
30 |
PDB header:signaling protein
Chain: P: PDB Molecule:insulin receptor;
PDBTitle: insulin receptor ectodomain in complex with two insulin molecules - c12 symmetry
|
|
|
|
| 18 | d1rgoa2
|
|
 |
14.5 |
83 |
Fold:CCCH zinc finger
Superfamily:CCCH zinc finger
Family:CCCH zinc finger |
|
|
|
| 19 | c6dftE_
|
|
 |
12.7 |
12 |
PDB header:transferase
Chain: E: PDB Molecule:deoxyhypusine synthase;
PDBTitle: trypanosoma brucei deoxyhypusine synthase
|
|
|
|
| 20 | d1x9na1
|
|
 |
12.0 |
13 |
Fold:ATP-dependent DNA ligase DNA-binding domain
Superfamily:ATP-dependent DNA ligase DNA-binding domain
Family:ATP-dependent DNA ligase DNA-binding domain |
|
|
|
| 21 | d1p3ie_ |
|
not modelled |
9.4 |
27 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
|
|
| 22 | d1id3a_ |
|
not modelled |
9.3 |
20 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
|
|
| 23 | d2pxyd2 |
|
not modelled |
9.0 |
44 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 24 | d1d5mb2 |
|
not modelled |
8.9 |
57 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 25 | c5mquA_ |
|
not modelled |
8.7 |
33 |
PDB header:virus
Chain: A: PDB Molecule:vp1;
PDBTitle: crystal structure of bovine enterovirus 2 determined with serial2 femtosecond x-ray crystallography
|
|
|
| 26 | d1klub2 |
|
not modelled |
8.6 |
57 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 27 | c2klqA_ |
|
not modelled |
8.6 |
75 |
PDB header:replication
Chain: A: PDB Molecule:dna replication licensing factor mcm6;
PDBTitle: the solution structure of cbd of human mcm6
|
|
|
| 28 | c4wm8A_ |
|
not modelled |
8.3 |
42 |
PDB header:virus
Chain: A: PDB Molecule:vp1;
PDBTitle: crystal structure of human enterovirus d68
|
|
|
| 29 | c5zyoD_ |
|
not modelled |
8.3 |
44 |
PDB header:transferase
Chain: D: PDB Molecule:ribosomal rna large subunit methyltransferase h;
PDBTitle: crystal structure of domain-swapped circular-permuted ybea (cp74) from2 escherichia coli
|
|
|
| 30 | c6iijA_ |
|
not modelled |
8.0 |
40 |
PDB header:viral protein
Chain: A: PDB Molecule:vp1;
PDBTitle: cryo-em structure of cv-a10 mature virion
|
|
|
| 31 | d1hxs1_ |
|
not modelled |
7.8 |
42 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 32 | d1pvc1_ |
|
not modelled |
7.8 |
42 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 33 | d2fmme1 |
|
not modelled |
7.6 |
21 |
Fold:ENT-like
Superfamily:ENT-like
Family:Emsy N terminal (ENT) domain-like |
|
|
| 34 | d1h8ta_ |
|
not modelled |
7.6 |
50 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 35 | c5lc5m_ |
|
not modelled |
7.4 |
13 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:nadh-ubiquinone oxidoreductase chain 4;
PDBTitle: structure of mammalian respiratory complex i, class2
|
|
|
| 36 | d1bx2b2 |
|
not modelled |
7.4 |
57 |
Fold:MHC antigen-recognition domain
Superfamily:MHC antigen-recognition domain
Family:MHC antigen-recognition domain |
|
|
| 37 | d1bev1_ |
|
not modelled |
7.3 |
38 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 38 | d1d4m1_ |
|
not modelled |
7.2 |
58 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 39 | d1cov1_ |
|
not modelled |
7.0 |
42 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 40 | d1dhsa_ |
|
not modelled |
6.9 |
13 |
Fold:DHS-like NAD/FAD-binding domain
Superfamily:DHS-like NAD/FAD-binding domain
Family:Deoxyhypusine synthase, DHS |
|
|
| 41 | c2fmmE_ |
|
not modelled |
6.8 |
21 |
PDB header:transcription
Chain: E: PDB Molecule:protein emsy;
PDBTitle: crystal structure of emsy-hp1 complex
|
|
|
| 42 | d1eqzg_ |
|
not modelled |
6.8 |
27 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
|
|
| 43 | d1eah1_ |
|
not modelled |
6.7 |
42 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 44 | c3mp7B_ |
|
not modelled |
6.6 |
40 |
PDB header:protein transport
Chain: B: PDB Molecule:preprotein translocase subunit sece;
PDBTitle: lateral opening of a translocon upon entry of protein suggests the2 mechanism of insertion into membranes
|
|
|
| 45 | c1z7s1_ |
|
not modelled |
6.4 |
42 |
PDB header:virus
Chain: 1: PDB Molecule:human coxsackievirus a21;
PDB Fragment:viral protein 1;
PDBTitle: the crystal structure of coxsackievirus a21
|
|
|
| 46 | d1y14b2 |
|
not modelled |
6.3 |
67 |
Fold:Dodecin subunit-like
Superfamily:N-terminal, heterodimerisation domain of RBP7 (RpoE)
Family:N-terminal, heterodimerisation domain of RBP7 (RpoE) |
|
|
| 47 | d2fdbm1 |
|
not modelled |
6.3 |
42 |
Fold:beta-Trefoil
Superfamily:Cytokine
Family:Fibroblast growth factors (FGF) |
|
|
| 48 | d1ev11_ |
|
not modelled |
6.2 |
50 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 49 | c4bipA_ |
|
not modelled |
6.2 |
46 |
PDB header:virus
Chain: A: PDB Molecule:vp1;
PDBTitle: homology model of coxsackievirus a7 (cav7) full capsid proteins.
|
|
|
| 50 | d1aym1_ |
|
not modelled |
6.1 |
50 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 51 | c1p58F_ |
|
not modelled |
5.9 |
36 |
PDB header:virus
Chain: F: PDB Molecule:envelope protein m;
PDBTitle: complex organization of dengue virus membrane proteins as revealed by2 9.5 angstrom cryo-em reconstruction
|
|
|
| 52 | d1kx5a_ |
|
not modelled |
5.9 |
24 |
Fold:Histone-fold
Superfamily:Histone-fold
Family:Nucleosome core histones |
|
|
| 53 | d1r1a1_ |
|
not modelled |
5.9 |
50 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 54 | c2xp1A_ |
|
not modelled |
5.8 |
25 |
PDB header:transcription
Chain: A: PDB Molecule:spt6;
PDBTitle: structure of the tandem sh2 domains from antonospora locustae2 transcription elongation factor spt6
|
|
|
| 55 | c3vbuA_ |
|
not modelled |
5.7 |
27 |
PDB header:virus
Chain: A: PDB Molecule:genome polyprotein, capsid protein vp1;
PDBTitle: crystal structure of empty human enterovirus 71 particle
|
|
|
| 56 | c3vbfA_ |
|
not modelled |
5.6 |
29 |
PDB header:virus
Chain: A: PDB Molecule:genome polyprotein, capsid protein vp1;
PDBTitle: crystal structure of formaldehyde treated human enterovirus 71 (space2 group i23)
|
|
|
| 57 | d1pov1_ |
|
not modelled |
5.6 |
42 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 58 | c5k0uA_ |
|
not modelled |
5.6 |
40 |
PDB header:virus
Chain: A: PDB Molecule:capsid protein vp1;
PDBTitle: cryoem structure of the full virion of a human rhinovirus c
|
|
|
| 59 | c5xtdo_ |
|
not modelled |
5.6 |
16 |
PDB header:oxidoreductase/electron transport
Chain: O: PDB Molecule:nadh dehydrogenase [ubiquinone] flavoprotein 2,
PDBTitle: cryo-em structure of human respiratory complex i
|
|
|
| 60 | c4q2gA_ |
|
not modelled |
5.5 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:phosphatidate cytidylyltransferase;
PDBTitle: crystal structure of an intramembrane cdp-dag synthetase central for2 phospholipid biosynthesis (s200c/s223c, inactive mutant)
|
|
|
| 61 | c5xs5A_ |
|
not modelled |
5.3 |
46 |
PDB header:virus
Chain: A: PDB Molecule:genome polyprotein;
PDBTitle: structure of coxsackievirus a6 (cva6) virus procapsid particle
|
|
|
| 62 | d1rhi1_ |
|
not modelled |
5.3 |
27 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 63 | d1mqta_ |
|
not modelled |
5.2 |
50 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 64 | d1oopa_ |
|
not modelled |
5.1 |
50 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 65 | d1ncqa_ |
|
not modelled |
5.1 |
27 |
Fold:Nucleoplasmin-like/VP (viral coat and capsid proteins)
Superfamily:Positive stranded ssRNA viruses
Family:Picornaviridae-like VP (VP1, VP2, VP3 and VP4) |
|
|
| 66 | c5jzgA_ |
|
not modelled |
5.0 |
40 |
PDB header:virus
Chain: A: PDB Molecule:capsid protein vp1;
PDBTitle: cryoem structure of the native empty particle of a human rhinovirus c
|
|
|