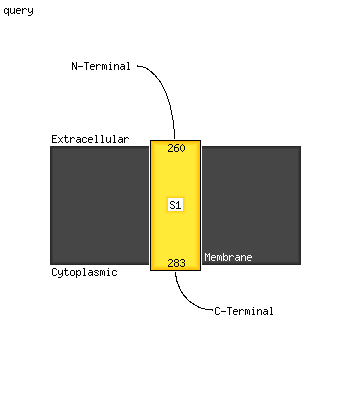
1 c5xfsB_
100.0
37
PDB header: protein transportChain: B: PDB Molecule: ppe family protein ppe15;PDBTitle: crystal structure of pe8-ppe15 in complex with espg5 from m.2 tuberculosis
2 d2g38b1
100.0
30
Fold: Ferritin-likeSuperfamily: PE/PPE dimer-likeFamily: PPE
3 c2g38B_
100.0
30
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: ppe family protein;PDBTitle: a pe/ppe protein complex from mycobacterium tuberculosis
4 c4xy3A_
100.0
15
PDB header: protein transportChain: A: PDB Molecule: esx-1 secretion-associated protein espb;PDBTitle: structure of esx-1 secreted protein espb
5 c4wj2A_
98.0
22
PDB header: unknown functionChain: A: PDB Molecule: antigen mtb48;PDBTitle: mycobacterial protein
6 c2vs0B_
97.8
11
PDB header: cell invasionChain: B: PDB Molecule: virulence factor esxa;PDBTitle: structural analysis of homodimeric staphylococcal aureus2 virulence factor esxa
7 c4iogD_
97.6
13
PDB header: unknown functionChain: D: PDB Molecule: secreted protein esxb;PDBTitle: the crystal structure of a secreted protein esxb (wild-type, in p212 space group) from bacillus anthracis str. sterne
8 c3gvmA_
97.6
16
PDB header: viral proteinChain: A: PDB Molecule: putative uncharacterized protein sag1039;PDBTitle: structure of the homodimeric wxg-100 family protein from streptococcus2 agalactiae
9 c3zbhC_
97.3
13
PDB header: unknown functionChain: C: PDB Molecule: esxa;PDBTitle: geobacillus thermodenitrificans esxa crystal form i
10 d1wa8a1
96.9
10
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
11 c4lwsB_
95.2
13
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
12 c4lwsA_
94.6
13
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein;PDBTitle: esxa : esxb (semet) hetero-dimer from thermomonospora curvata
13 d1wa8b1
94.6
17
Fold: Ferritin-likeSuperfamily: EsxAB dimer-likeFamily: ESAT-6 like
14 c4i0xA_
93.6
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: esat-6-like protein mab_3112;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
15 c2kg7B_
89.2
20
PDB header: unknown functionChain: B: PDB Molecule: esat-6-like protein esxh;PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
16 c4i0xJ_
67.6
16
PDB header: structural genomics, unknown functionChain: J: PDB Molecule: esat-6-like protein mab_3113;PDBTitle: crystal structure of the mycobacterum abscessus esxef (mab_3112-2 mab_3113) complex
17 d1ui5a2
58.9
20
Fold: Tetracyclin repressor-like, C-terminal domainSuperfamily: Tetracyclin repressor-like, C-terminal domainFamily: Tetracyclin repressor-like, C-terminal domain
18 c3zfsA_
27.6
28
PDB header: oxidoreductaseChain: A: PDB Molecule: f420-reducing hydrogenase, subunit alpha;PDBTitle: cryo-em structure of the f420-reducing nife-hydrogenase from a2 methanogenic archaeon with bound substrate
19 d1dlpa1
17.2
18
Fold: beta-Prism IISuperfamily: alpha-D-mannose-specific plant lectinsFamily: alpha-D-mannose-specific plant lectins
20 d1zeea1
16.4
42
Fold: Indolic compounds 2,3-dioxygenase-likeSuperfamily: Indolic compounds 2,3-dioxygenase-likeFamily: Indoleamine 2,3-dioxygenase-like
21 c4xb6D_
not modelled
12.4
17
PDB header: transferaseChain: D: PDB Molecule: alpha-d-ribose 1-methylphosphonate 5-phosphate c-p lyase;PDBTitle: structure of the e. coli c-p lyase core complex
22 c1bkvA_
not modelled
12.1
50
PDB header: structural proteinChain: A: PDB Molecule: t3-785;PDBTitle: collagen
23 c2kg7A_
not modelled
11.7
28
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized protein esxg (pe family protein);PDBTitle: structure and features of the complex formed by the tuberculosis2 virulence factors rv0287 and rv0288
24 c1bkvC_
not modelled
11.6
50
PDB header: structural proteinChain: C: PDB Molecule: t3-785;PDBTitle: collagen
25 c1bkvB_
not modelled
11.6
50
PDB header: structural proteinChain: B: PDB Molecule: t3-785;PDBTitle: collagen
26 c2nvjA_
not modelled
10.9
40
PDB header: hydrolaseChain: A: PDB Molecule: 25mer peptide from vacuolar atp synthase subunitPDBTitle: nmr structures of transmembrane segment from subunit a from2 the yeast proton v-atpase
27 d1vlfn1
not modelled
9.7
100
Fold: Prealbumin-likeSuperfamily: Cna protein B-type domainFamily: Cna protein B-type domain
28 c5abvF_
not modelled
8.9
45
PDB header: translationChain: F: PDB Molecule: gh11071p;PDBTitle: complex of d. melanogaster eif4e with the 4e-binding2 protein mextli
29 c5l85B_
not modelled
8.1
36
PDB header: signaling proteinChain: B: PDB Molecule: nuclear fragile x mental retardation-interacting protein 1;PDBTitle: solution structure of the complex between human znhit3 and nufip12 proteins
30 c6aokA_
not modelled
7.9
25
PDB header: hydrolaseChain: A: PDB Molecule: ceg4;PDBTitle: crystal structure of legionella pneumophila effector ceg4 with n-2 terminal tev protease cleavage sequence
31 c6nbiP_
not modelled
7.9
60
PDB header: signaling proteinChain: P: PDB Molecule: long-acting parathyroid hormone analog;PDBTitle: cryo-em structure of parathyroid hormone receptor type 1 in complex2 with a long-acting parathyroid hormone analog and g protein
32 c1bzgA_
not modelled
7.7
29
PDB header: hormoneChain: A: PDB Molecule: parathyroid hormone-related protein;PDBTitle: the solution structure of human parathyroid hormone-related2 protein (1-34) in near-physiological solution, nmr, 303 structures
33 c2iu1A_
not modelled
7.3
28
PDB header: transcriptionChain: A: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: crystal structure of eif5 c-terminal domain
34 c6cgjA_
not modelled
7.2
38
PDB header: hydrolaseChain: A: PDB Molecule: effector protein lem4 (lpg1101);PDBTitle: structure of the had domain of effector protein lem4 (lpg1101) from2 legionella pneumophila
35 c3sjrB_
not modelled
7.0
13
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of conserved unkown function protein cv_1783 from2 chromobacterium violaceum atcc 12472
36 c2fulE_
not modelled
6.6
28
PDB header: translationChain: E: PDB Molecule: eukaryotic translation initiation factor 5;PDBTitle: crystal structure of the c-terminal domain of s. cerevisiae eif5
37 c1vytF_
not modelled
6.3
75
PDB header: transport proteinChain: F: PDB Molecule: voltage-dependent l-type calcium channelPDBTitle: beta3 subunit complexed with aid
38 d1fcda3
not modelled
5.9
23
Fold: CO dehydrogenase flavoprotein C-domain-likeSuperfamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domainFamily: FAD/NAD-linked reductases, dimerisation (C-terminal) domain
39 c2jtwA_
not modelled
5.4
50
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane helix 7 of yeast vatpase;PDBTitle: solution structure of tm7 bound to dpc micelles
40 c4u39Q_
not modelled
5.4
36
PDB header: cell cycleChain: Q: PDB Molecule: cell division factor;PDBTitle: crystal structure of ftsz:mciz complex from bacillus subtilis
41 c5hl8B_
not modelled
5.3
20
PDB header: protein transportChain: B: PDB Molecule: type ii secretion system protein l;PDBTitle: 1.93 angstrom resolution crystal structure of a pullulanase-specific2 type ii secretion system integral cytoplasmic membrane protein gspl3 (c-terminal fragment; residues 309-397) from klebsiella pneumoniae4 subsp. pneumoniae ntuh-k2044
42 c2lkqA_
not modelled
5.3
56
PDB header: immune systemChain: A: PDB Molecule: immunoglobulin lambda-like polypeptide 1;PDBTitle: nmr structure of the lambda 5 22-45 peptide
43 c4yv4D_
not modelled
5.2
33
PDB header: structural proteinChain: D: PDB Molecule: spindle assembly abnormal protein 5;PDBTitle: structure of the c. elegans sas-5 coiled coil domain
44 c5bv9A_
not modelled
5.2
50
PDB header: hydrolaseChain: A: PDB Molecule: cellulose 1,4-beta-cellobiosidase;PDBTitle: the structure of bacillus pumilus gh48 in complex with cellobiose