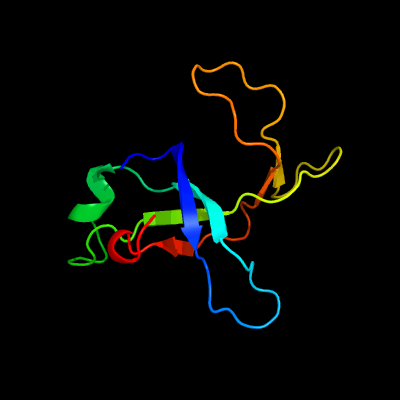
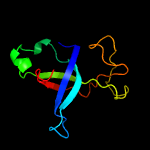
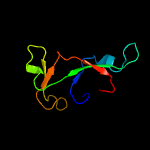
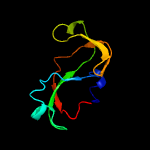
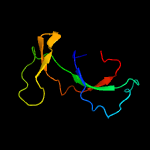
1 d1m1fa_
100.0
33
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
2 c5hjzA_
100.0
60
PDB header: hydrolase/rnaChain: A: PDB Molecule: endoribonuclease mazf9;PDBTitle: structure of m. tuberculosis mazf-mt1 (rv2801c) in complex with rna
3 c4mzpC_
100.0
25
PDB header: hydrolaseChain: C: PDB Molecule: mazf mrna interferase;PDBTitle: mazf from s. aureus crystal form iii, c2221, 2.7 a
4 d1ne8a_
99.9
24
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
5 d1ub4a_
99.9
30
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: Kid/PemK
6 c5hk3B_
99.9
29
PDB header: hydrolase/dnaChain: B: PDB Molecule: endoribonuclease mazf6;PDBTitle: crystal structure of m. tuberculosis mazf-mt3 t52d-f62d mutant in2 complex with dna
7 c5xe3B_
99.9
32
PDB header: hydrolase/antitoxinChain: B: PDB Molecule: endoribonuclease mazf4;PDBTitle: endoribonuclease in complex with its cognate antitoxin from2 mycobacterial species
8 c5wygC_
99.9
33
PDB header: hydrolaseChain: C: PDB Molecule: probable endoribonuclease mazf7;PDBTitle: the crystal structure of the apo form of mtb mazf
9 c5ccaA_
99.8
17
PDB header: hydrolaseChain: A: PDB Molecule: endoribonuclease mazf3;PDBTitle: crystal structure of mtb toxin
10 c3jrzA_
89.2
16
PDB header: toxinChain: A: PDB Molecule: ccdb;PDBTitle: ccdbvfi-formii-ph5.6
11 d3vuba_
66.1
14
Fold: SH3-like barrelSuperfamily: Cell growth inhibitor/plasmid maintenance toxic componentFamily: CcdB
12 c5ikjA_
59.8
38
PDB header: transcriptionChain: A: PDB Molecule: cryptic loci regulator 2;PDBTitle: structure of clr2 bound to the clr1 c-terminus
13 c3mxuA_
41.5
29
PDB header: oxidoreductaseChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of glycine cleavage system protein h from bartonella2 henselae
14 c6n1bA_
39.3
16
PDB header: hydrolaseChain: A: PDB Molecule: carbohydrate-binding protein;PDBTitle: crystal structure of an n-acetylgalactosamine deacetylase from f.2 plautii in complex with blood group b trisaccharide
15 c2edgA_
35.1
17
PDB header: biosynthetic proteinChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: solution structure of the gcv_h domain from mouse glycine
16 d1onla_
30.3
25
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains
17 d1okja2
29.8
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: YeaZ-like
18 c3a8jF_
26.8
25
PDB header: transferase/transport proteinChain: F: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of et-ehred complex
19 c2vhmI_
26.4
14
PDB header: ribosomeChain: I: PDB Molecule: 50s ribosomal protein l11;PDBTitle: structure of pdf binding helix in complex with the ribosome2 (part 1 of 4)
20 d1hpca_
22.8
29
Fold: Barrel-sandwich hybridSuperfamily: Single hybrid motifFamily: Biotinyl/lipoyl-carrier proteins and domains
21 c3cjqB_
not modelled
22.6
14
PDB header: transferase/ribosomal proteinChain: B: PDB Molecule: 50s ribosomal protein l11;PDBTitle: ribosomal protein l11 methyltransferase (prma) in complex with2 dimethylated ribosomal protein l11 in space group p212121
22 c3iftA_
not modelled
22.0
29
PDB header: oxidoreductaseChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of glycine cleavage system protein h from2 mycobacterium tuberculosis, using x-rays from the compact light3 source.
23 c2ftcG_
not modelled
21.5
11
PDB header: ribosomeChain: G: PDB Molecule: 39s ribosomal protein l11, mitochondrial;PDBTitle: structural model for the large subunit of the mammalian mitochondrial2 ribosome
24 c3j21H_
not modelled
21.3
19
PDB header: ribosomeChain: H: PDB Molecule: 50s ribosomal protein l11p;PDBTitle: promiscuous behavior of proteins in archaeal ribosomes revealed by2 cryo-em: implications for evolution of eukaryotic ribosomes (50s3 ribosomal proteins)
25 d1ex0a1
not modelled
17.9
21
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: Transglutaminase N-terminal domain
26 c1vw4V_
not modelled
16.4
15
PDB header: ribosomeChain: V: PDB Molecule: 54s ribosomal protein l36, mitochondrial;PDBTitle: structure of the yeast mitochondrial large ribosomal subunit
27 c1l1kA_
not modelled
15.9
14
PDB header: protein bindingChain: A: PDB Molecule: barstar;PDBTitle: nmr identification and characterization of the flexible2 regions in the 160 kd molten globule-like aggregate of3 barstar at low ph
28 d2daqa1
not modelled
13.4
21
Fold: SH3-like barrelSuperfamily: Tudor/PWWP/MBTFamily: PWWP domain
29 c2fqpD_
not modelled
11.9
14
PDB header: metal binding proteinChain: D: PDB Molecule: hypothetical protein bp2299;PDBTitle: crystal structure of a cupin domain (bp2299) from bordetella pertussis2 tohama i at 1.80 a resolution
30 c1cffB_
not modelled
10.5
30
PDB header: calmodulinChain: B: PDB Molecule: calcium pump;PDBTitle: nmr solution structure of a complex of calmodulin with a binding2 peptide of the ca2+-pump
31 c1jqmA_
not modelled
9.8
23
PDB header: ribosomeChain: A: PDB Molecule: 50s ribosomal protein l11;PDBTitle: fitting of l11 protein and elongation factor g (ef-g) in2 the cryo-em map of e. coli 70s ribosome bound with ef-g,3 gdp and fusidic acid
32 c1vowJ_
not modelled
8.7
13
PDB header: ribosomeChain: J: PDB Molecule: 50s ribosomal protein l11;PDBTitle: crystal structure of five 70s ribosomes from escherichia coli in2 complex with protein y. this file contains the 50s subunit of one 70s3 ribosome. the entire crystal structure contains five 70s ribosomes4 and is described in remark 400.
33 c4mnoA_
not modelled
8.5
18
PDB header: translationChain: A: PDB Molecule: translation initiation factor 1a;PDBTitle: crystal structure of aif1a from pyrococcus abyssi
34 c3d55A_
not modelled
8.5
31
PDB header: toxin inhibitorChain: A: PDB Molecule: uncharacterized protein rv3357/mt3465;PDBTitle: crystal structure of m. tuberculosis yefm antitoxin
35 c5o60J_
not modelled
8.1
20
PDB header: ribosomeChain: J: PDB Molecule: 50s ribosomal protein l11;PDBTitle: structure of the 50s large ribosomal subunit from mycobacterium2 smegmatis
36 c4p7xA_
not modelled
7.9
29
PDB header: oxidoreductaseChain: A: PDB Molecule: l-proline cis-4-hydroxylase;PDBTitle: l-pipecolic acid-bound l-proline cis-4-hydroxylase
37 c5a35A_
not modelled
7.8
9
PDB header: transport proteinChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: crystal structure of glycine cleavage protein h-like (gcvh-l)2 from streptococcus pyogenes
38 c2jysA_
not modelled
7.6
0
PDB header: hydrolaseChain: A: PDB Molecule: protease/reverse transcriptase;PDBTitle: solution structure of simian foamy virus (mac) protease
39 d2gycg2
not modelled
7.5
15
Fold: Ribosomal L11/L12e N-terminal domainSuperfamily: Ribosomal L11/L12e N-terminal domainFamily: Ribosomal L11/L12e N-terminal domain
40 c2wseE_
not modelled
7.4
17
PDB header: photosynthesisChain: E: PDB Molecule: photosystem i reaction center subunit iv a, chloroplastic;PDBTitle: improved model of plant photosystem i
41 c3bboK_
not modelled
7.2
24
PDB header: ribosomeChain: K: PDB Molecule: ribosomal protein l11;PDBTitle: homology model for the spinach chloroplast 50s subunit fitted to 9.4a2 cryo-em map of the 70s chlororibosome
42 c4rmoA_
not modelled
7.1
24
PDB header: toxin/rnaChain: A: PDB Molecule: cptn toxin;PDBTitle: crystal structure of the cptin type iii toxin-antitoxin system from2 eubacterium rectale
43 c3oeiB_
not modelled
7.1
31
PDB header: toxin, protein bindingChain: B: PDB Molecule: relj (antitoxin rv3357);PDBTitle: crystal structure of mycobacterium tuberculosis reljk (rv3357-rv3358-2 relbe3)
44 d1zaka2
not modelled
6.2
21
Fold: Rubredoxin-likeSuperfamily: Microbial and mitochondrial ADK, insert "zinc finger" domainFamily: Microbial and mitochondrial ADK, insert "zinc finger" domain
45 c3cezA_
not modelled
6.1
19
PDB header: oxidoreductaseChain: A: PDB Molecule: methionine-r-sulfoxide reductase;PDBTitle: crystal structure of methionine-r-sulfoxide reductase from2 burkholderia pseudomallei
46 c2ka7A_
not modelled
6.0
25
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: glycine cleavage system h protein;PDBTitle: nmr solution structure of tm0212 at 40 c
47 c1s1iK_
not modelled
5.7
11
PDB header: ribosomeChain: K: PDB Molecule: 60s ribosomal protein l12;PDBTitle: structure of the ribosomal 80s-eef2-sordarin complex from yeast2 obtained by docking atomic models for rna and protein components into3 a 11.7 a cryo-em map. this file, 1s1i, contains 60s subunit. the 40s4 ribosomal subunit is in file 1s1h.
48 c2dgyA_
not modelled
5.6
40
PDB header: translationChain: A: PDB Molecule: mgc11102 protein;PDBTitle: solution structure of the eukaryotic initiation factor 1a2 in mgc11102 protein
49 c5cuzA_
not modelled
5.4
29
PDB header: chaperoneChain: A: PDB Molecule: methylmalonic aciduria and homocystinuria type d protein,PDBTitle: crystal structure of semet-substituted n-terminal truncated human b12-2 chaperone cbld (108-296)