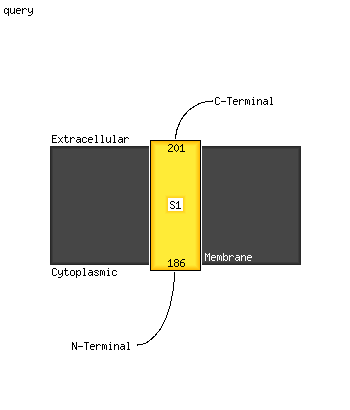
1 c2vhiG_
100.0
18
PDB header: hydrolaseChain: G: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
2 c1emsB_
100.0
28
PDB header: antitumor proteinChain: B: PDB Molecule: nit-fragile histidine triad fusion protein;PDBTitle: crystal structure of the c. elegans nitfhit protein
3 c2vhhA_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: cg3027-pa;PDBTitle: crystal structure of a pyrimidine degrading enzyme from2 drosophila melanogaster
4 c2w1vA_
100.0
33
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase homolog 2;PDBTitle: crystal structure of mouse nitrilase-2 at 1.4a resolution
5 c6ftqA_
100.0
18
PDB header: hydrolaseChain: A: PDB Molecule: beta-ureidopropionase;PDBTitle: crystal structure of human beta-ureidopropionase (beta-alanine2 synthase) - mutant t299c
6 c5h8lM_
100.0
21
PDB header: hydrolaseChain: M: PDB Molecule: n-carbamoylputrescine amidohydrolase;PDBTitle: crystal structure of medicago truncatula n-carbamoylputrescine2 amidohydrolase (mtcpa) c158s mutant in complex with putrescine
7 d1uf5a_
100.0
24
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase
8 c2plqA_
100.0
21
PDB header: hydrolaseChain: A: PDB Molecule: aliphatic amidase;PDBTitle: crystal structure of the amidase from geobacillus pallidus rapc8
9 c2e2kC_
100.0
17
PDB header: hydrolaseChain: C: PDB Molecule: formamidase;PDBTitle: helicobacter pylori formamidase amif contains a fine-tuned cysteine-2 glutamate-lysine catalytic triad
10 c4hg3C_
100.0
28
PDB header: hydrolaseChain: C: PDB Molecule: probable hydrolase nit2;PDBTitle: structural insights into yeast nit2: wild-type yeast nit2 in complex2 with alpha-ketoglutarate
11 c2e11B_
100.0
29
PDB header: hydrolaseChain: B: PDB Molecule: hydrolase;PDBTitle: the crystal structure of xc1258 from xanthomonas campestris: a cn-2 hydrolase superfamily protein with an arsenic adduct in the active3 site
12 c6mg6D_
100.0
19
PDB header: hydrolaseChain: D: PDB Molecule: carbon-nitrogen hydrolase;PDBTitle: crystal structure of carbon-nitrogen hydrolase from helicobacter2 pylori g27
13 d1emsa2
100.0
28
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase
14 c5khaA_
100.0
21
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad+ synthetase;PDBTitle: structure of glutamine-dependent nad+ synthetase from acinetobacter2 baumannii in complex with adenosine diphosphate (adp)
15 c3dlaD_
100.0
21
PDB header: ligaseChain: D: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: x-ray crystal structure of glutamine-dependent nad+ synthetase from2 mycobacterium tuberculosis bound to naad+ and don
16 d1f89a_
100.0
25
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Nitrilase
17 c3wuyA_
100.0
22
PDB header: hydrolaseChain: A: PDB Molecule: nitrilase;PDBTitle: crystal structure of nit6803
18 c4f4hA_
100.0
23
PDB header: ligaseChain: A: PDB Molecule: glutamine dependent nad+ synthetase;PDBTitle: crystal structure of a glutamine dependent nad+ synthetase from2 burkholderia thailandensis
19 d1j31a_
100.0
26
Fold: Carbon-nitrogen hydrolaseSuperfamily: Carbon-nitrogen hydrolaseFamily: Carbamilase
20 c3n05B_
100.0
25
PDB header: ligaseChain: B: PDB Molecule: nh(3)-dependent nad(+) synthetase;PDBTitle: crystal structure of nh3-dependent nad+ synthetase from streptomyces2 avermitilis
21 c3hkxA_
not modelled
100.0
30
PDB header: hydrolaseChain: A: PDB Molecule: amidase;PDBTitle: crystal structure analysis of an amidase from nesterenkonia sp.
22 c3ilvA_
not modelled
100.0
16
PDB header: ligaseChain: A: PDB Molecule: glutamine-dependent nad(+) synthetase;PDBTitle: crystal structure of a glutamine-dependent nad(+) synthetase2 from cytophaga hutchinsonii
23 c4cyyA_
not modelled
100.0
20
PDB header: hydrolaseChain: A: PDB Molecule: pantetheinase;PDBTitle: the structure of vanin-1: defining the link between metabolic disease,2 oxidative stress and inflammation
24 c5vrhA_
not modelled
100.0
16
PDB header: transferaseChain: A: PDB Molecule: apolipoprotein n-acyltransferase;PDBTitle: apolipoprotein n-acyltransferase c387s active site mutant
25 c5n6mA_
not modelled
100.0
20
PDB header: membrane proteinChain: A: PDB Molecule: apolipoprotein n-acyltransferase;PDBTitle: structure of the membrane integral lipoprotein n-acyltransferase lnt2 from p. aeruginosa
26 c5hyyA_
not modelled
100.0
16
PDB header: hydrolaseChain: A: PDB Molecule: nta1p;PDBTitle: crystal structure of n-terminal amidase
27 c2csuB_
not modelled
78.2
9
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: 457aa long hypothetical protein;PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
28 d1s3la_
not modelled
74.1
18
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: YfcE-like
29 c1s3mA_
not modelled
73.3
18
PDB header: phosphodiesteraseChain: A: PDB Molecule: hypothetical protein mj0936;PDBTitle: structural and functional characterization of a novel2 archaeal phosphodiesterase
30 c3qfnA_
not modelled
70.9
21
PDB header: hydrolaseChain: A: PDB Molecule: putative uncharacterized protein;PDBTitle: crystal structure of streptococcal asymmetric ap4a hydrolase and2 phosphodiesterase spr1479/saph in complex with inorganic phosphate
31 d1uf3a_
not modelled
68.8
22
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: TT1561-like
32 d1i60a_
not modelled
60.9
11
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: IolI-like
33 c1su1A_
not modelled
59.9
27
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yfce;PDBTitle: structural and biochemical characterization of yfce, a phosphoesterase2 from e. coli
34 d1su1a_
not modelled
59.9
27
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: YfcE-like
35 d1o60a_
not modelled
58.1
10
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase
36 c6ncsB_
not modelled
56.0
18
PDB header: biosynthetic proteinChain: B: PDB Molecule: n-acetylneuraminic acid (sialic acid) synthetase;PDBTitle: crystal structure of n-acetylneuraminic acid (sialic acid) synthetase2 from leptospira borgpetersenii serovar hardjo-bovis in complex with3 citrate
37 d1d9ea_
not modelled
54.7
9
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: Class I DAHP synthetase
38 d1t70a_
not modelled
46.8
16
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: DR1281-like
39 d1gsaa1
not modelled
42.9
11
Fold: PreATP-grasp domainSuperfamily: PreATP-grasp domainFamily: Prokaryotic glutathione synthetase, N-terminal domain
40 c5zfsA_
not modelled
42.4
17
PDB header: isomeraseChain: A: PDB Molecule: d-allulose-3-epimerase;PDBTitle: crystal structure of arthrobacter globiformis m30 sugar epimerase2 which can produce d-allulose from d-fructose
41 c3stgA_
not modelled
40.6
14
PDB header: transferaseChain: A: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase;PDBTitle: crystal structure of a58p, del(n59), and loop 7 truncated mutant of 3-2 deoxy-d-manno-octulosonate 8-phosphate synthase (kdo8ps) from3 neisseria meningitidis
42 c4lu0A_
not modelled
39.6
12
PDB header: transferaseChain: A: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase;PDBTitle: crystal structure of 2-keto-3-deoxy-d-manno-octulosonate-8-phosphate2 synthase from pseudomonas aeruginosa.
43 c2fu3A_
not modelled
36.8
11
PDB header: biosynthetic protein/structural proteinChain: A: PDB Molecule: gephyrin;PDBTitle: crystal structure of gephyrin e-domain
44 c4k3zA_
not modelled
36.5
13
PDB header: oxidoreductaseChain: A: PDB Molecule: d-erythrulose 4-phosphate dehydrogenase;PDBTitle: crystal structure of d-erythrulose 4-phosphate dehydrogenase from2 brucella melitensis, solved by iodide sad
45 c3dx5A_
not modelled
36.0
9
PDB header: lyaseChain: A: PDB Molecule: uncharacterized protein asbf;PDBTitle: crystal structure of the probable 3-dhs dehydratase asbf involved in2 the petrobactin synthesis from bacillus anthracis
46 c2ou4C_
not modelled
34.8
14
PDB header: isomeraseChain: C: PDB Molecule: d-tagatose 3-epimerase;PDBTitle: crystal structure of d-tagatose 3-epimerase from2 pseudomonas cichorii
47 c3vniC_
not modelled
34.5
6
PDB header: isomeraseChain: C: PDB Molecule: xylose isomerase domain protein tim barrel;PDBTitle: crystal structures of d-psicose 3-epimerase from clostridium2 cellulolyticum h10 and its complex with ketohexose sugars
48 c4r8kC_
not modelled
33.4
11
PDB header: hydrolaseChain: C: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of the guinea pig l-asparaginase 1 catalytic domain
49 c2qw5B_
not modelled
33.0
10
PDB header: isomeraseChain: B: PDB Molecule: xylose isomerase-like tim barrel;PDBTitle: crystal structure of a putative sugar phosphate isomerase/epimerase2 (ava4194) from anabaena variabilis atcc 29413 at 1.78 a resolution
50 c4yajA_
not modelled
32.8
15
PDB header: ligaseChain: A: PDB Molecule: alpha subunit of acetyl-coenzyme a synthetasePDBTitle: ca. korarchaeum cryptofilum dinucleotide forming acetyl-coenzyme a2 synthetase 1 (apo form)
51 c3wqoB_
not modelled
30.1
10
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein mj1311;PDBTitle: crystal structure of d-tagatose 3-epimerase-like protein
52 c2ei9A_
not modelled
29.2
23
PDB header: gene regulationChain: A: PDB Molecule: non-ltr retrotransposon r1bmks orf2 protein;PDBTitle: crystal structure of r1bm endonuclease domain
53 d4pgaa_
not modelled
29.1
13
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase
54 d1t71a_
not modelled
28.6
15
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: DR1281-like
55 c2ejcA_
not modelled
28.6
9
PDB header: ligaseChain: A: PDB Molecule: pantoate--beta-alanine ligase;PDBTitle: crystal structure of pantoate--beta-alanine ligase (panc)2 from thermotoga maritima
56 d1agxa_
not modelled
28.5
13
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase
57 d2zdra2
not modelled
28.1
16
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like
58 c2p2dA_
not modelled
27.8
11
PDB header: transferaseChain: A: PDB Molecule: l-asparaginase i;PDBTitle: crystal structure and allosteric regulation of the cytoplasmic2 escherichia coli l-asparaginase i
59 d1v8fa_
not modelled
27.0
21
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)
60 d1ihoa_
not modelled
26.9
18
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Nucleotidylyl transferaseFamily: Pantothenate synthetase (Pantoate-beta-alanine ligase, PanC)
61 d2yvta1
not modelled
26.7
18
Fold: Metallo-dependent phosphatasesSuperfamily: Metallo-dependent phosphatasesFamily: TT1561-like
62 c5hmqE_
not modelled
26.2
15
PDB header: lyaseChain: E: PDB Molecule: 4-hydroxyphenylpyruvate dioxygenase;PDBTitle: xylose isomerase-like tim barrel/4-hydroxyphenylpyruvate dioxygenase2 fusion protein
63 c3mxtA_
not modelled
25.3
15
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine ligase from campylobacter2 jejuni
64 c3uk2B_
not modelled
25.3
9
PDB header: ligaseChain: B: PDB Molecule: pantothenate synthetase;PDBTitle: the structure of pantothenate synthetase from burkholderia2 thailandensis
65 c3vylB_
not modelled
25.2
14
PDB header: isomeraseChain: B: PDB Molecule: l-ribulose 3-epimerase;PDBTitle: structure of l-ribulose 3-epimerase
66 c1gshA_
not modelled
25.1
11
PDB header: glutathione biosynthesis ligaseChain: A: PDB Molecule: glutathione biosynthetic ligase;PDBTitle: structure of escherichia coli glutathione synthetase at ph 7.5
67 d2vgna3
not modelled
24.6
11
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like
68 c2zdsB_
not modelled
24.5
14
PDB header: dna binding proteinChain: B: PDB Molecule: putative dna-binding protein;PDBTitle: crystal structure of sco6571 from streptomyces coelicolor a3(2)
69 c3o3cD_
not modelled
24.3
13
PDB header: transferaseChain: D: PDB Molecule: glycogen [starch] synthase isoform 2;PDBTitle: glycogen synthase basal state udp complex
70 c2nqqA_
not modelled
24.3
10
PDB header: biosynthetic proteinChain: A: PDB Molecule: molybdopterin biosynthesis protein moea;PDBTitle: moea r137q
71 c3qxbB_
not modelled
23.9
11
PDB header: isomeraseChain: B: PDB Molecule: putative xylose isomerase;PDBTitle: crystal structure of a putative xylose isomerase (yp_426450.1) from2 rhodospirillum rubrum atcc 11170 at 1.90 a resolution
72 c3guzB_
not modelled
23.0
18
PDB header: ligaseChain: B: PDB Molecule: pantothenate synthetase;PDBTitle: structural and substrate-binding studies of pantothenate2 synthenate (ps)provide insights into homotropic inhibition3 by pantoate in ps's
73 c3rqzC_
not modelled
22.7
29
PDB header: hydrolaseChain: C: PDB Molecule: metallophosphoesterase;PDBTitle: crystal structure of metallophosphoesterase from sphaerobacter2 thermophilus
74 c5g2rA_
not modelled
22.2
14
PDB header: transferaseChain: A: PDB Molecule: molybdopterin biosynthesis protein cnx1;PDBTitle: crystal structure of the mo-insertase domain cnx1e from2 arabidopsis thaliana
75 c3n8hA_
not modelled
22.1
12
PDB header: ligaseChain: A: PDB Molecule: pantothenate synthetase;PDBTitle: crystal structure of pantoate-beta-alanine ligase from francisella2 tularensis
76 c3aytA_
not modelled
21.8
15
PDB header: unknown functionChain: A: PDB Molecule: putative uncharacterized protein tthb071;PDBTitle: tthb071 protein from thermus thermophilus hb8
77 d2a40b1
not modelled
21.1
17
Fold: DNase I-likeSuperfamily: DNase I-likeFamily: DNase I-like
78 d2ocda1
not modelled
21.1
8
Fold: Glutaminase/AsparaginaseSuperfamily: Glutaminase/AsparaginaseFamily: Glutaminase/Asparaginase
79 c4f1hA_
not modelled
20.9
10
PDB header: hydrolase/dnaChain: A: PDB Molecule: tyrosyl-dna phosphodiesterase 2;PDBTitle: crystal structure of tdp2 from danio rerio complexed with a single2 strand dna
80 d1yx1a1
not modelled
20.8
12
Fold: TIM beta/alpha-barrelSuperfamily: Xylose isomerase-likeFamily: KguE-like
81 c3sz8D_
not modelled
20.8
14
PDB header: transferaseChain: D: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase 2;PDBTitle: crystal structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 burkholderia pseudomallei
82 c3tebA_
not modelled
20.4
10
PDB header: hydrolaseChain: A: PDB Molecule: endonuclease/exonuclease/phosphatase;PDBTitle: endonuclease/exonuclease/phosphatase family protein from leptotrichia2 buccalis c-1013-b
83 c6bngB_
not modelled
20.4
10
PDB header: transferaseChain: B: PDB Molecule: 2-dehydro-3-deoxyphosphooctonate aldolase;PDBTitle: structure of 2-dehydro-3-deoxyphosphooctonate aldolase from2 acinetobacter baumannii
84 c3pnxF_
not modelled
20.3
8
PDB header: transferaseChain: F: PDB Molecule: putative sulfurtransferase dsre;PDBTitle: crystal structure of a putative sulfurtransferase dsre (swol_2425)2 from syntrophomonas wolfei str. goettingen at 1.92 a resolution
85 c2o14A_
not modelled
20.3
11
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: hypothetical protein yxim;PDBTitle: x-ray crystal structure of protein yxim_bacsu from bacillus subtilis.2 northeast structural genomics consortium target sr595
86 c5d95A_
not modelled
19.5
19
PDB header: transferaseChain: A: PDB Molecule: aminotransferase class-iii;PDBTitle: structure of thermostable omega-transaminase
87 c2nydB_
not modelled
19.4
12
PDB header: unknown functionChain: B: PDB Molecule: upf0135 protein sa1388;PDBTitle: crystal structure of staphylococcus aureus hypothetical protein sa1388
88 c1xuzA_
not modelled
18.4
18
PDB header: biosynthetic proteinChain: A: PDB Molecule: polysialic acid capsule biosynthesis protein siac;PDBTitle: crystal structure analysis of sialic acid synthase (neub)from2 neisseria meningitidis, bound to mn2+, phosphoenolpyruvate, and n-3 acetyl mannosaminitol
89 d1vlia2
not modelled
17.5
15
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: NeuB-like
90 c2odrC_
not modelled
16.9
20
PDB header: ligaseChain: C: PDB Molecule: phosphoseryl-trna synthetase;PDBTitle: methanococcus maripaludis phosphoseryl-trna synthetase
91 c4s1aB_
not modelled
16.7
11
PDB header: unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a hypothetical protein cthe_0052 from2 ruminiclostridium thermocellum atcc 27405
92 c1vliA_
not modelled
16.4
17
PDB header: biosynthetic proteinChain: A: PDB Molecule: spore coat polysaccharide biosynthesis protein spse;PDBTitle: crystal structure of spore coat polysaccharide biosynthesis protein2 spse (bsu37870) from bacillus subtilis at 2.38 a resolution
93 c3ju2A_
not modelled
15.7
17
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: uncharacterized protein smc04130;PDBTitle: crystal structure of protein smc04130 from sinorhizobium meliloti 1021
94 c2issF_
not modelled
15.7
10
PDB header: lyase, transferaseChain: F: PDB Molecule: glutamine amidotransferase subunit pdxt;PDBTitle: structure of the plp synthase holoenzyme from thermotoga maritima
95 c2odrB_
not modelled
15.6
14
PDB header: ligaseChain: B: PDB Molecule: phosphoseryl-trna synthetase;PDBTitle: methanococcus maripaludis phosphoseryl-trna synthetase
96 c5tnvA_
not modelled
15.5
16
PDB header: isomeraseChain: A: PDB Molecule: ap endonuclease, family protein 2;PDBTitle: crystal structure of a xylose isomerase-like tim barrel protein from2 mycobacterium smegmatis in complex with magnesium
97 c2zsmA_
not modelled
15.3
21
PDB header: isomeraseChain: A: PDB Molecule: glutamate-1-semialdehyde 2,1-aminomutase;PDBTitle: crystal structure of glutamate-1-semialdehyde 2,1-2 aminomutase from aeropyrum pernix, hexagonal form
98 d1x52a1
not modelled
15.2
7
Fold: Bacillus chorismate mutase-likeSuperfamily: L30e-likeFamily: ERF1/Dom34 C-terminal domain-like
99 c3cnyA_
not modelled
15.1
15
PDB header: biosynthetic proteinChain: A: PDB Molecule: inositol catabolism protein iole;PDBTitle: crystal structure of a putative inositol catabolism protein iole2 (iole, lp_3607) from lactobacillus plantarum wcfs1 at 1.85 a3 resolution