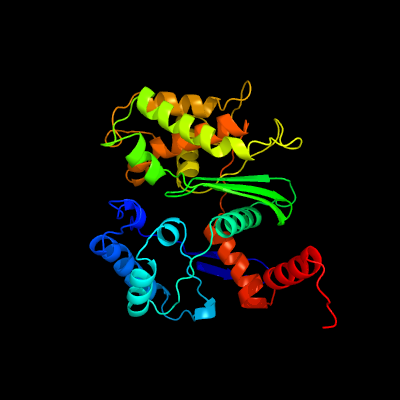
| 1 | c2floA_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:exopolyphosphatase;
PDBTitle: crystal structure of exopolyphosphatase (ppx) from e. coli o157:h7
|
|
|
|
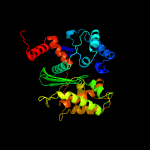
| 2 | c3cerD_
|
|
 |
100.0 |
22 |
PDB header:structural genomics, unknown function
Chain: D: PDB Molecule:possible exopolyphosphatase-like protein;
PDBTitle: crystal structure of the exopolyphosphatase-like protein q8g5j2.2 northeast structural genomics consortium target blr13
|
|
|
|
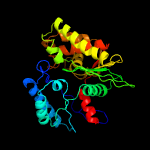
| 3 | c3mdqA_
|
|
 |
100.0 |
23 |
PDB header:hydrolase
Chain: A: PDB Molecule:exopolyphosphatase;
PDBTitle: crystal structure of an exopolyphosphatase (chu_0316) from cytophaga2 hutchinsonii atcc 33406 at 1.50 a resolution
|
|
|
|
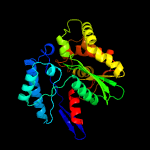
| 4 | c3hi0B_
|
|
 |
100.0 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:putative exopolyphosphatase;
PDBTitle: crystal structure of putative exopolyphosphatase (17739545) from2 agrobacterium tumefaciens str. c58 (dupont) at 2.30 a resolution
|
|
|
|
| 5 | c1t6dB_
|
|
 |
100.0 |
25 |
PDB header:hydrolase
Chain: B: PDB Molecule:exopolyphosphatase;
PDBTitle: miras phasing of the aquifex aeolicus ppx/gppa phosphatase: crystal2 structure of the type ii variant
|
|
|
|
| 6 | d1t6ca2
|
|
 |
100.0 |
24 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ppx/GppA phosphatase |
|
|
|
| 7 | d1u6za3
|
|
 |
100.0 |
22 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ppx/GppA phosphatase |
|
|
|
| 8 | d1t6ca1
|
|
 |
100.0 |
23 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ppx/GppA phosphatase |
|
|
|
| 9 | d1u6za2
|
|
 |
100.0 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ppx/GppA phosphatase |
|
|
|
| 10 | c3aapA_
|
|
 |
99.8 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleoside triphosphate diphosphohydrolase i;
PDBTitle: crystal structure of lp1ntpdase from legionella pneumophila
|
|
|
|
| 11 | c5u7xF_
|
|
 |
99.7 |
17 |
PDB header:hydrolase
Chain: F: PDB Molecule:nod factor binding lectin-nucleotide phosphohydrolase;
PDBTitle: crystal structure of a nucleoside triphosphate diphosphohydrolase2 (ntpdase) from the legume vigna unguiculata subsp. cylindrica3 (dolichos biflorus) in complex with phosphate and manganese
|
|
|
|
| 12 | c5u7wA_
|
|
 |
99.7 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:apyrase;
PDBTitle: crystal structure of a nucleoside triphosphate diphosphohydrolase2 (ntpdase) from the legume trifolium repens in complex with adenine3 and phosphate
|
|
|
|
| 13 | c3zx2A_
|
|
 |
99.7 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleoside triphosphate diphosphohydrolase 1;
PDBTitle: ntpdase1 in complex with decavanadate
|
|
|
|
| 14 | c3cj9A_
|
|
 |
99.3 |
17 |
PDB header:hydrolase
Chain: A: PDB Molecule:ectonucleoside triphosphate diphosphohydrolase 2;
PDBTitle: structure of rattus norvegicus ntpdase2 in complex with2 calcium, amp and phosphate
|
|
|
|
| 15 | c3agrB_
|
|
 |
98.7 |
20 |
PDB header:hydrolase
Chain: B: PDB Molecule:nucleoside triphosphate hydrolase;
PDBTitle: crystal structure of nucleoside triphosphate hydrolases from neospora2 caninum
|
|
|
|
| 16 | c3h1qB_
|
|
 |
98.7 |
15 |
PDB header:structural protein
Chain: B: PDB Molecule:ethanolamine utilization protein eutj;
PDBTitle: crystal structure of ethanolamine utilization protein eutj from2 carboxydothermus hydrogenoformans
|
|
|
|
| 17 | c4a5bA_
|
|
 |
98.6 |
22 |
PDB header:hydrolase
Chain: A: PDB Molecule:nucleoside-triphosphatase 2;
PDBTitle: crystal structure of the c258s/c268s variant of toxoplasma gondii2 nucleoside triphosphate diphosphohydrolase 1 (ntpdase1)
|
|
|
|
| 18 | c5eoxB_
|
|
 |
98.2 |
13 |
PDB header:peptide binding protein
Chain: B: PDB Molecule:type 4 fimbrial biogenesis protein pilm;
PDBTitle: pseudomonas aeruginosa pilm bound to adp
|
|
|
|
| 19 | c1e4gT_
|
|
 |
98.1 |
10 |
PDB header:bacterial cell division
Chain: T: PDB Molecule:cell division protein ftsa;
PDBTitle: ftsa (atp-bound form) from thermotoga maritima
|
|
|
|
| 20 | c4czeA_
|
|
 |
98.0 |
16 |
PDB header:structural protein
Chain: A: PDB Molecule:rod shape-determining protein mreb;
PDBTitle: c. crescentus mreb, double filament, empty
|
|
|
|
| 21 | c2ychA_ |
|
not modelled |
98.0 |
17 |
PDB header:cell cycle
Chain: A: PDB Molecule:competence protein pilm;
PDBTitle: pilm-piln type iv pilus biogenesis complex
|
|
|
| 22 | c3wqtB_ |
|
not modelled |
98.0 |
11 |
PDB header:structural genomics
Chain: B: PDB Molecule:cell division protein ftsa;
PDBTitle: staphylococcus aureus ftsa complexed with amppnp
|
|
|
| 23 | c3js6A_ |
|
not modelled |
97.9 |
12 |
PDB header:unknown function
Chain: A: PDB Molecule:uncharacterized parm protein;
PDBTitle: crystal structure of apo psk41 parm protein
|
|
|
| 24 | c4apwH_ |
|
not modelled |
97.8 |
12 |
PDB header:structural protein
Chain: H: PDB Molecule:alp12;
PDBTitle: alp12 filament structure
|
|
|
| 25 | c2d0oA_ |
|
not modelled |
97.7 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:diol dehydratase-reactivating factor large
PDBTitle: strcuture of diol dehydratase-reactivating factor complexed2 with adp and mg2+
|
|
|
| 26 | c1mwmA_ |
|
not modelled |
97.7 |
14 |
PDB header:structural protein
Chain: A: PDB Molecule:parm;
PDBTitle: parm from plasmid r1 adp form
|
|
|
| 27 | d1e4ft2 |
|
not modelled |
97.5 |
9 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 28 | c5tkyA_ |
|
not modelled |
97.4 |
12 |
PDB header:chaperone
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of the co-translational hsp70 chaperone ssb in the2 atp-bound, open conformation
|
|
|
| 29 | c5mb9B_ |
|
not modelled |
97.4 |
14 |
PDB header:chaperone
Chain: B: PDB Molecule:putative heat shock protein;
PDBTitle: crystal structure of the eukaryotic ribosome associated complex (rac),2 a unique hsp70/hsp40 pair
|
|
|
| 30 | d2zgya2 |
|
not modelled |
97.4 |
17 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 31 | c2fsnB_ |
|
not modelled |
97.3 |
16 |
PDB header:structural protein
Chain: B: PDB Molecule:hypothetical protein ta0583;
PDBTitle: crystal structure of ta0583, an archaeal actin homolog, complex with2 adp
|
|
|
| 32 | d1jcea2 |
|
not modelled |
97.1 |
17 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 33 | c5obuA_ |
|
not modelled |
97.1 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein dnak;
PDBTitle: mycoplasma genitalium dnak deletion mutant lacking sbdalpha in complex2 with amppnp.
|
|
|
| 34 | c4xe7A_ |
|
not modelled |
97.1 |
11 |
PDB header:structural protein
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: bacillus thuringiensis parm in apo form
|
|
|
| 35 | d2fsja1 |
|
not modelled |
97.1 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Ta0583-like |
|
|
| 36 | c3d2fC_ |
|
not modelled |
96.8 |
12 |
PDB header:chaperone
Chain: C: PDB Molecule:heat shock protein homolog sse1;
PDBTitle: crystal structure of a complex of sse1p and hsp70
|
|
|
| 37 | c3dwlB_ |
|
not modelled |
96.8 |
10 |
PDB header:structural protein
Chain: B: PDB Molecule:actin-related protein 3;
PDBTitle: crystal structure of fission yeast arp2/3 complex lacking the arp22 subunit
|
|
|
| 38 | c6gfaA_ |
|
not modelled |
96.8 |
13 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein 105 kda;
PDBTitle: structure of nucleotide binding domain of hsp110, atp and mg2+2 complexed
|
|
|
| 39 | c1jcgA_ |
|
not modelled |
96.8 |
12 |
PDB header:structural protein
Chain: A: PDB Molecule:rod shape-determining protein mreb;
PDBTitle: mreb from thermotoga maritima, amppnp
|
|
|
| 40 | c2v7yA_ |
|
not modelled |
96.7 |
16 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein dnak;
PDBTitle: crystal structure of the molecular chaperone dnak from2 geobacillus kaustophilus hta426 in post-atp hydrolysis3 state
|
|
|
| 41 | c4gniA_ |
|
not modelled |
96.7 |
13 |
PDB header:chaperone
Chain: A: PDB Molecule:putative heat shock protein;
PDBTitle: structure of the ssz1 atpase bound to atp and magnesium
|
|
|
| 42 | c5jygA_ |
|
not modelled |
96.7 |
14 |
PDB header:structural protein
Chain: A: PDB Molecule:actin-like atpase;
PDBTitle: cryo-em structure of the mamk filament at 6.5 a
|
|
|
| 43 | c3c7nB_ |
|
not modelled |
96.6 |
16 |
PDB header:chaperone/chaperone
Chain: B: PDB Molecule:heat shock cognate;
PDBTitle: structure of the hsp110:hsc70 nucleotide exchange complex
|
|
|
| 44 | c4j8fA_ |
|
not modelled |
96.6 |
18 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock 70 kda protein 1a/1b, hsc70-interacting protein;
PDBTitle: crystal structure of a fusion protein containing the nbd of hsp70 and2 the middle domain of hip
|
|
|
| 45 | c6izrK_ |
|
not modelled |
96.5 |
8 |
PDB header:protein fibril
Chain: K: PDB Molecule:putative plasmid segregation protein parm;
PDBTitle: whole structure of a 15-stranded parm filament from clostridium2 botulinum
|
|
|
| 46 | c2khoA_ |
|
not modelled |
96.2 |
15 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock protein 70;
PDBTitle: nmr-rdc / xray structure of e. coli hsp70 (dnak) chaperone (1-605)2 complexed with adp and substrate
|
|
|
| 47 | c5e84B_ |
|
not modelled |
96.1 |
14 |
PDB header:chaperone
Chain: B: PDB Molecule:78 kda glucose-regulated protein;
PDBTitle: atp-bound state of bip
|
|
|
| 48 | c4xhpA_ |
|
not modelled |
96.1 |
18 |
PDB header:structural protein
Chain: A: PDB Molecule:parm hybrid fusion protein;
PDBTitle: bacillus thuringiensis parm hybrid protein with adp, containing two2 parm mutants
|
|
|
| 49 | c1dkgD_ |
|
not modelled |
95.9 |
16 |
PDB header:complex (hsp24/hsp70)
Chain: D: PDB Molecule:molecular chaperone dnak;
PDBTitle: crystal structure of the nucleotide exchange factor grpe2 bound to the atpase domain of the molecular chaperone dnak
|
|
|
| 50 | c4pl7B_ |
|
not modelled |
95.8 |
15 |
PDB header:structural protein, contractile protein
Chain: B: PDB Molecule:actin,thymosin beta-4;
PDBTitle: structure of komagataella pastoris actin-thymosin beta4 hybrid
|
|
|
| 51 | c1o1f4_ |
|
not modelled |
95.6 |
14 |
PDB header:contractile protein
Chain: 4: PDB Molecule:skeletal muscle actin;
PDBTitle: molecular models of averaged rigor crossbridges from tomograms of2 insect flight muscle
|
|
|
| 52 | c5afuB_ |
|
not modelled |
95.3 |
13 |
PDB header:motor protein
Chain: B: PDB Molecule:dynactin;
PDBTitle: cryo-em structure of dynein tail-dynactin-bicd2n complex
|
|
|
| 53 | c3uleB_ |
|
not modelled |
95.0 |
17 |
PDB header:structural protein
Chain: B: PDB Molecule:actin-related protein 2;
PDBTitle: structure of bos taurus arp2/3 complex with bound inhibitor ck-869 and2 atp
|
|
|
| 54 | c4jd2B_ |
|
not modelled |
95.0 |
9 |
PDB header:structural protein
Chain: B: PDB Molecule:actin-related protein 2;
PDBTitle: crystal structure of bos taurus arp2/3 complex binding with mus2 musculus gmf
|
|
|
| 55 | d1dkgd2 |
|
not modelled |
95.0 |
23 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 56 | c2v7zA_ |
|
not modelled |
94.8 |
12 |
PDB header:chaperone
Chain: A: PDB Molecule:heat shock cognate 71 kda protein;
PDBTitle: crystal structure of the 70-kda heat shock cognate protein2 from rattus norvegicus in post-atp hydrolysis state
|
|
|
| 57 | c3qb0C_ |
|
not modelled |
94.3 |
15 |
PDB header:structural protein
Chain: C: PDB Molecule:actin-related protein 4;
PDBTitle: crystal structure of actin-related protein arp4 from s. cerevisiae2 complexed with atp
|
|
|
| 58 | c2p9lA_ |
|
not modelled |
94.3 |
13 |
PDB header:structural protein
Chain: A: PDB Molecule:actin-like protein 3;
PDBTitle: crystal structure of bovine arp2/3 complex
|
|
|
| 59 | d1bupa2 |
|
not modelled |
94.2 |
28 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 60 | c6etxH_ |
|
not modelled |
94.2 |
10 |
PDB header:dna binding protein
Chain: H: PDB Molecule:actin-related protein 5;
PDBTitle: cryo-em structure of the human ino80 complex bound to nucleosome
|
|
|
| 61 | d2e8aa2 |
|
not modelled |
94.0 |
32 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 62 | c6bqwH_ |
|
not modelled |
93.8 |
11 |
PDB header:cytosolic protein
Chain: H: PDB Molecule:bacterial actin alfa;
PDBTitle: alfa filament bound to amppnp
|
|
|
| 63 | c4kboA_ |
|
not modelled |
93.3 |
11 |
PDB header:signaling protein
Chain: A: PDB Molecule:stress-70 protein, mitochondrial;
PDBTitle: crystal structure of the human mortalin (grp75) atpase domain in the2 apo form
|
|
|
| 64 | c3iucC_ |
|
not modelled |
92.9 |
17 |
PDB header:chaperone
Chain: C: PDB Molecule:heat shock 70kda protein 5 (glucose-regulated
PDBTitle: crystal structure of the human 70kda heat shock protein 52 (bip/grp78) atpase domain in complex with adp
|
|
|
| 65 | d1nm1a2 |
|
not modelled |
92.7 |
21 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 66 | c6gejR_ |
|
not modelled |
92.1 |
13 |
PDB header:nuclear protein
Chain: R: PDB Molecule:actin-like protein arp6;
PDBTitle: chromatin remodeller-nucleosome complex at 3.6 a resolution.
|
|
|
| 67 | d2hf3a2 |
|
not modelled |
91.9 |
21 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 68 | c4fo0A_ |
|
not modelled |
91.8 |
21 |
PDB header:gene regulation
Chain: A: PDB Molecule:actin-related protein 8;
PDBTitle: human actin-related protein arp8 in its atp-bound state
|
|
|
| 69 | c6fhsJ_ |
|
not modelled |
91.6 |
12 |
PDB header:dna binding protein
Chain: J: PDB Molecule:arp5;
PDBTitle: cryoem structure of ino80core
|
|
|
| 70 | c2p9kB_ |
|
not modelled |
91.4 |
17 |
PDB header:structural protein
Chain: B: PDB Molecule:actin-like protein 2;
PDBTitle: crystal structure of bovine arp2/3 complex co-crystallized with atp2 and crosslinked with glutaraldehyde
|
|
|
| 71 | d2fxua2 |
|
not modelled |
91.4 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 72 | c4i6mA_ |
|
not modelled |
91.4 |
13 |
PDB header:transcription/hydrolase
Chain: A: PDB Molecule:actin-related protein 7;
PDBTitle: structure of arp7-arp9-snf2(hsa)-rtt102 subcomplex of swi/snf2 chromatin remodeler.
|
|
|
| 73 | c4i6mB_ |
|
not modelled |
91.2 |
15 |
PDB header:transcription/hydrolase
Chain: B: PDB Molecule:actin-like protein arp9;
PDBTitle: structure of arp7-arp9-snf2(hsa)-rtt102 subcomplex of swi/snf2 chromatin remodeler.
|
|
|
| 74 | c5aftJ_ |
|
not modelled |
91.0 |
21 |
PDB header:motor protein
Chain: J: PDB Molecule:actin related protein 11;
PDBTitle: cryoem structure of dynactin complex at 4.0 angstrom2 resolution
|
|
|
| 75 | c1hpmA_ |
|
not modelled |
91.0 |
14 |
PDB header:hydrolase (acting on acid anhydrides)
Chain: A: PDB Molecule:44k atpase fragment (n-terminal) of 7o kd heat-
PDBTitle: how potassium affects the activity of the molecular2 chaperone hsc70. ii. potassium binds specifically in the3 atpase active site
|
|
|
| 76 | c1nbwA_ |
|
not modelled |
90.9 |
19 |
PDB header:hydrolase
Chain: A: PDB Molecule:glycerol dehydratase reactivase alpha subunit;
PDBTitle: glycerol dehydratase reactivase
|
|
|
| 77 | d1k8kb1 |
|
not modelled |
90.7 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 78 | c4cj7B_ |
|
not modelled |
90.3 |
14 |
PDB header:structural protein
Chain: B: PDB Molecule:actin/actin family protein;
PDBTitle: structure of crenactin, an archeal actin-like protein
|
|
|
| 79 | d1yaga2 |
|
not modelled |
90.2 |
22 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 80 | c3bfjK_ |
|
not modelled |
89.5 |
17 |
PDB header:oxidoreductase
Chain: K: PDB Molecule:1,3-propanediol oxidoreductase;
PDBTitle: crystal structure analysis of 1,3-propanediol oxidoreductase
|
|
|
| 81 | c6fmlJ_ |
|
not modelled |
88.7 |
12 |
PDB header:dna binding protein
Chain: J: PDB Molecule:actin related protein 5;
PDBTitle: cryoem structure ino80core nucleosome complex
|
|
|
| 82 | d1k8ka2 |
|
not modelled |
87.3 |
15 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Actin/HSP70 |
|
|
| 83 | c4rtfD_ |
|
not modelled |
87.2 |
14 |
PDB header:chaperone
Chain: D: PDB Molecule:chaperone protein dnak;
PDBTitle: crystal structure of molecular chaperone dnak from mycobacterium2 tuberculosis h37rv
|
|
|
| 84 | c4fr2A_ |
|
not modelled |
87.0 |
18 |
PDB header:oxidoreductase, metal binding protein
Chain: A: PDB Molecule:1,3-propanediol dehydrogenase;
PDBTitle: alcohol dehydrogenase from oenococcus oeni
|
|
|
| 85 | c5l9wb_ |
|
not modelled |
85.8 |
19 |
PDB header:ligase
Chain: B: PDB Molecule:acetophenone carboxylase gamma subunit;
PDBTitle: crystal structure of the apc core complex
|
|
|
| 86 | c4am6A_ |
|
not modelled |
83.8 |
19 |
PDB header:nuclear protein
Chain: A: PDB Molecule:actin-like protein arp8;
PDBTitle: c-terminal domain of actin-related protein arp8 from s. cerevisiae
|
|
|
| 87 | c5yvmA_ |
|
not modelled |
83.2 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: crystal structure of the archaeal halo-thermophilic red sea brine pool2 alcohol dehydrogenase adh/d1 bound to nzq
|
|
|
| 88 | c3ox4D_ |
|
not modelled |
81.3 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:alcohol dehydrogenase 2;
PDBTitle: structures of iron-dependent alcohol dehydrogenase 2 from zymomonas2 mobilis zm4 complexed with nad cofactor
|
|
|
| 89 | c1ta9A_ |
|
not modelled |
80.0 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol dehydrogenase;
PDBTitle: crystal structure of glycerol dehydrogenase from schizosaccharomyces2 pombe
|
|
|
| 90 | c3ce9A_ |
|
not modelled |
79.8 |
11 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:glycerol dehydrogenase;
PDBTitle: crystal structure of glycerol dehydrogenase (np_348253.1) from2 clostridium acetobutylicum at 2.37 a resolution
|
|
|
| 91 | c5m45K_ |
|
not modelled |
79.6 |
28 |
PDB header:ligase
Chain: K: PDB Molecule:acetone carboxylase beta subunit;
PDBTitle: structure of acetone carboxylase purified from xanthobacter2 autotrophicus
|
|
|
| 92 | c3cetA_ |
|
not modelled |
77.3 |
13 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:conserved archaeal protein;
PDBTitle: crystal structure of the pantheonate kinase-like protein q6m145 at the2 resolution 1.8 a. northeast structural genomics consortium target3 mrr63
|
|
|
| 93 | d1saza1 |
|
not modelled |
74.8 |
19 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Acetokinase-like |
|
|
| 94 | c2dpnB_ |
|
not modelled |
73.3 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of the glycerol kinase from thermus2 thermophilus hb8
|
|
|
| 95 | c5hvnA_ |
|
not modelled |
73.2 |
15 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: 3.0 angstrom crystal structure of 3-dehydroquinate synthase (arob)2 from francisella tularensis in complex with nad.
|
|
|
| 96 | d1rrma_ |
|
not modelled |
73.1 |
13 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 97 | c3g25B_ |
|
not modelled |
71.3 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:glycerol kinase;
PDBTitle: 1.9 angstrom crystal structure of glycerol kinase (glpk) from2 staphylococcus aureus in complex with glycerol.
|
|
|
| 98 | d1o2da_ |
|
not modelled |
70.4 |
13 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 99 | c3flcX_ |
|
not modelled |
67.7 |
7 |
PDB header:transferase
Chain: X: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of the his-tagged h232r mutant of glycerol kinase2 from enterococcus casseliflavus with glycerol
|
|
|
| 100 | c6c76A_ |
|
not modelled |
66.3 |
19 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:alcohol dehydrogenase;
PDBTitle: structure of iron containing alcohol dehydrogenase from thermococcus2 thioreducens in an orthorhombic crystal form
|
|
|
| 101 | c4mcaB_ |
|
not modelled |
64.8 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:glycerol dehydrogenase;
PDBTitle: crystal structure of glycerol dehydrogenase from serratia to 1.9a
|
|
|
| 102 | c5ya2A_ |
|
not modelled |
64.8 |
15 |
PDB header:structural protein
Chain: A: PDB Molecule:autoinducer-2 kinase;
PDBTitle: crystal structure of lsrk-hpr complex with adp
|
|
|
| 103 | d1zc6a1 |
|
not modelled |
64.4 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:BadF/BadG/BcrA/BcrD-like |
|
|
| 104 | c5l9wB_ |
|
not modelled |
63.9 |
18 |
PDB header:ligase
Chain: B: PDB Molecule:acetophenone carboxylase gamma subunit;
PDBTitle: crystal structure of the apc core complex
|
|
|
| 105 | c2d4wA_ |
|
not modelled |
62.1 |
12 |
PDB header:transferase
Chain: A: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of glycerol kinase from cellulomonas sp.2 nt3060
|
|
|
| 106 | d2p3ra1 |
|
not modelled |
60.8 |
14 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Glycerol kinase |
|
|
| 107 | d2d0oa3 |
|
not modelled |
60.4 |
29 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ATPase domain of dehydratase reactivase alpha subunit |
|
|
| 108 | d1jq5a_ |
|
not modelled |
59.9 |
14 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 109 | c6jkpD_ |
|
not modelled |
59.8 |
14 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:methanol dehydrogenase;
PDBTitle: crystal structure of sulfoacetaldehyde reductase from bifidobacterium2 kashiwanohense in complex with nad+
|
|
|
| 110 | c3zokB_ |
|
not modelled |
59.4 |
15 |
PDB header:lyase
Chain: B: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: structure of 3-dehydroquinate synthase from actinidia chinensis in2 complex with nad
|
|
|
| 111 | d2i1qa1 |
|
not modelled |
58.1 |
27 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
|
|
| 112 | d1nbwa3 |
|
not modelled |
58.1 |
30 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:ATPase domain of dehydratase reactivase alpha subunit |
|
|
| 113 | d1szpa1 |
|
not modelled |
56.5 |
20 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
|
|
| 114 | d1kq3a_ |
|
not modelled |
55.2 |
17 |
Fold:Dehydroquinate synthase-like
Superfamily:Dehydroquinate synthase-like
Family:Iron-containing alcohol dehydrogenase |
|
|
| 115 | c3ezwD_ |
|
not modelled |
54.6 |
13 |
PDB header:transferase
Chain: D: PDB Molecule:glycerol kinase;
PDBTitle: crystal structure of a hyperactive escherichia coli glycerol kinase2 mutant gly230 --> asp obtained using microfluidic crystallization3 devices
|
|
|
| 116 | c3okfA_ |
|
not modelled |
54.5 |
14 |
PDB header:lyase
Chain: A: PDB Molecule:3-dehydroquinate synthase;
PDBTitle: 2.5 angstrom resolution crystal structure of 3-dehydroquinate synthase2 (arob) from vibrio cholerae
|
|
|
| 117 | c3hl0B_ |
|
not modelled |
54.2 |
12 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:maleylacetate reductase;
PDBTitle: crystal structure of maleylacetate reductase from agrobacterium2 tumefaciens
|
|
|
| 118 | c3cqyA_ |
|
not modelled |
53.8 |
10 |
PDB header:transferase
Chain: A: PDB Molecule:anhydro-n-acetylmuramic acid kinase;
PDBTitle: crystal structure of a functionally unknown protein (so_1313) from2 shewanella oneidensis mr-1
|
|
|
| 119 | d1szpb1 |
|
not modelled |
52.1 |
20 |
Fold:SAM domain-like
Superfamily:Rad51 N-terminal domain-like
Family:DNA repair protein Rad51, N-terminal domain |
|
|
| 120 | c1xupO_ |
|
not modelled |
49.9 |
7 |
PDB header:transferase
Chain: O: PDB Molecule:glycerol kinase;
PDBTitle: enterococcus casseliflavus glycerol kinase complexed with glycerol
|
|
|