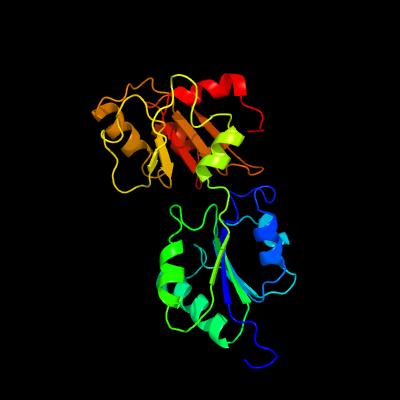
| 1 | c1pjtB_
|
|
 |
100.0 |
29 |
PDB header:transferase/oxidoreductase/lyase
Chain: B: PDB Molecule:siroheme synthase;
PDBTitle: the structure of the ser128ala point-mutant variant of cysg, the2 multifunctional methyltransferase/dehydrogenase/ferrochelatase for3 siroheme synthesis
|
|
|
|
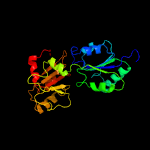
| 2 | d1s4da_
|
|
 |
100.0 |
28 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
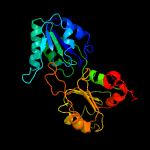
| 3 | c2yboA_
|
|
 |
100.0 |
31 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the x-ray structure of the sam-dependent uroporphyrinogen2 iii methyltransferase nire from pseudomonas aeruginosa in3 complex with sah
|
|
|
|
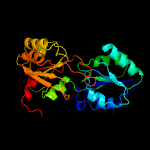
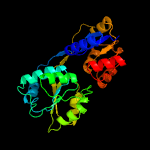
| 4 | c4es6A_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of hemd (pa5259) from pseudomonas aeruginosa (pao1)2 at 2.22 a resolution
|
|
|
|
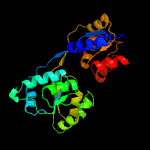
| 5 | c3re1B_
|
|
 |
100.0 |
22 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthetase;
PDBTitle: crystal structure of uroporphyrinogen iii synthase from pseudomonas2 syringae pv. tomato dc3000
|
|
|
|
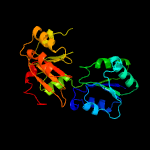
| 6 | d1pjqa2
|
|
 |
100.0 |
29 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
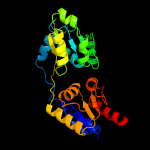
| 7 | d1wd7a_
|
|
 |
100.0 |
20 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
|
|
|
| 8 | d1jr2a_
|
|
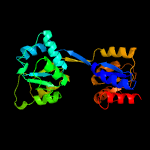 |
100.0 |
21 |
Fold:HemD-like
Superfamily:HemD-like
Family:HemD-like |
|
|
|
| 9 | c1jr2A_
|
|
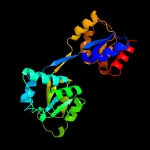 |
100.0 |
21 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: structure of uroporphyrinogen iii synthase
|
|
|
|
| 10 | d1cbfa_
|
|
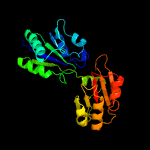 |
100.0 |
26 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 11 | c1cbfA_
|
|
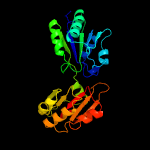 |
100.0 |
26 |
PDB header:methyltransferase
Chain: A: PDB Molecule:cobalt-precorrin-4 transmethylase;
PDBTitle: the x-ray structure of a cobalamin biosynthetic enzyme, cobalt2 precorrin-4 methyltransferase, cbif
|
|
|
|
| 12 | c3d8tB_
|
|
 |
100.0 |
21 |
PDB header:lyase
Chain: B: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: thermus thermophilus uroporphyrinogen iii synthase
|
|
|
|
| 13 | c4e16A_
|
|
 |
100.0 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: precorrin-4 c(11)-methyltransferase from clostridium difficile
|
|
|
|
| 14 | c3mw8A_
|
|
 |
100.0 |
23 |
PDB header:lyase
Chain: A: PDB Molecule:uroporphyrinogen-iii synthase;
PDBTitle: crystal structure of an uroporphyrinogen-iii synthase (sama_3255) from2 shewanella amazonensis sb2b at 1.65 a resolution
|
|
|
|
| 15 | c3kwpA_
|
|
 |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:predicted methyltransferase;
PDBTitle: crystal structure of putative methyltransferase from lactobacillus2 brevis
|
|
|
|
| 16 | d1ve2a1
|
|
 |
100.0 |
26 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 17 | c3ndcB_
|
|
 |
100.0 |
23 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-4 c(11)-methyltransferase;
PDBTitle: crystal structure of precorrin-4 c11-methyltransferase from2 rhodobacter capsulatus
|
|
|
|
| 18 | c5n0sA_
|
|
 |
100.0 |
17 |
PDB header:transferase
Chain: A: PDB Molecule:peptide n-methyltransferase;
PDBTitle: crystal structure of opha-deltac6 mutant y98a in complex with sam
|
|
|
|
| 19 | c5hw4C_
|
|
 |
100.0 |
25 |
PDB header:transferase
Chain: C: PDB Molecule:ribosomal rna small subunit methyltransferase i;
PDBTitle: crystal structure of escherichia coli 16s rrna methyltransferase rsmi2 in complex with adomet
|
|
|
|
| 20 | d1va0a1
|
|
 |
100.0 |
29 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
|
| 21 | c2zvbA_ |
|
not modelled |
100.0 |
25 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-3 c17-methyltransferase;
PDBTitle: crystal structure of tt0207 from thermus thermophilus hb8
|
|
|
| 22 | d1wyza1 |
|
not modelled |
100.0 |
12 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 23 | d1wdea_ |
|
not modelled |
100.0 |
20 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 24 | c3nutC_ |
|
not modelled |
100.0 |
24 |
PDB header:transferase
Chain: C: PDB Molecule:precorrin-3 methylase;
PDBTitle: crystal structure of the methyltransferase cobj
|
|
|
| 25 | d2deka1 |
|
not modelled |
100.0 |
22 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 26 | d1vhva_ |
|
not modelled |
100.0 |
16 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 27 | c2e0kA_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 c20-methyltransferase;
PDBTitle: crystal structure of cbil, a methyltransferase involved in anaerobic2 vitamin b12 biosynthesis
|
|
|
| 28 | c3i4tA_ |
|
not modelled |
100.0 |
16 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:diphthine synthase;
PDBTitle: crystal structure of putative diphthine synthase from entamoeba2 histolytica
|
|
|
| 29 | c2qbuA_ |
|
not modelled |
100.0 |
20 |
PDB header:transferase
Chain: A: PDB Molecule:precorrin-2 methyltransferase;
PDBTitle: crystal structure of methanothermobacter thermautotrophicus cbil
|
|
|
| 30 | c2npnA_ |
|
not modelled |
100.0 |
22 |
PDB header:transferase
Chain: A: PDB Molecule:putative cobalamin synthesis related protein;
PDBTitle: crystal structure of putative cobalamin synthesis related protein2 (cobf) from corynebacterium diphtheriae
|
|
|
| 31 | c3nd1B_ |
|
not modelled |
100.0 |
15 |
PDB header:transferase
Chain: B: PDB Molecule:precorrin-6a synthase/cobf protein;
PDBTitle: crystal structure of precorrin-6a synthase from rhodobacter capsulatus
|
|
|
| 32 | c3p9zA_ |
|
not modelled |
100.0 |
16 |
PDB header:ligase
Chain: A: PDB Molecule:uroporphyrinogen iii cosynthase (hemd);
PDBTitle: crystal structure of uroporphyrinogen-iii synthetase from helicobacter2 pylori 26695
|
|
|
| 33 | c2bb3B_ |
|
not modelled |
100.0 |
21 |
PDB header:transferase
Chain: B: PDB Molecule:cobalamin biosynthesis precorrin-6y methylase (cbie);
PDBTitle: crystal structure of cobalamin biosynthesis precorrin-6y methylase2 (cbie) from archaeoglobus fulgidus
|
|
|
| 34 | d2bb3a1 |
|
not modelled |
100.0 |
21 |
Fold:Tetrapyrrole methylase
Superfamily:Tetrapyrrole methylase
Family:Tetrapyrrole methylase |
|
|
| 35 | c3hh1D_ |
|
not modelled |
99.9 |
23 |
PDB header:transferase
Chain: D: PDB Molecule:tetrapyrrole methylase family protein;
PDBTitle: the structure of a tetrapyrrole methylase family protein domain from2 chlorobium tepidum tls
|
|
|
| 36 | c3fq6A_ |
|
not modelled |
99.2 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:methyltransferase;
PDBTitle: the crystal structure of a methyltransferase domain from bacteroides2 thetaiotaomicron vpi
|
|
|
| 37 | c2csuB_ |
|
not modelled |
97.5 |
11 |
PDB header:structural genomics, unknown function
Chain: B: PDB Molecule:457aa long hypothetical protein;
PDBTitle: crystal structure of ph0766 from pyrococcus horikoshii ot3
|
|
|
| 38 | c3d64A_ |
|
not modelled |
95.5 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 burkholderia pseudomallei
|
|
|
| 39 | c6f3oC_ |
|
not modelled |
95.4 |
15 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 pseudomonas aeruginosa complexed with adenine, k+ and zn2+ cations
|
|
|
| 40 | c6aphA_ |
|
not modelled |
95.4 |
16 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of adenosylhomocysteinase from elizabethkingia2 anophelis nuhp1 in complex with nad and adenosine
|
|
|
| 41 | c5butG_ |
|
not modelled |
95.2 |
13 |
PDB header:membrane protein
Chain: G: PDB Molecule:ktr system potassium uptake protein a,ktr system potassium
PDBTitle: crystal structure of inactive conformation of ktrab k+ transporter
|
|
|
| 42 | c2yxbA_ |
|
not modelled |
95.2 |
18 |
PDB header:isomerase
Chain: A: PDB Molecule:coenzyme b12-dependent mutase;
PDBTitle: crystal structure of the methylmalonyl-coa mutase alpha-subunit from2 aeropyrum pernix
|
|
|
| 43 | c4yajA_ |
|
not modelled |
95.1 |
12 |
PDB header:ligase
Chain: A: PDB Molecule:alpha subunit of acetyl-coenzyme a synthetase
PDBTitle: ca. korarchaeum cryptofilum dinucleotide forming acetyl-coenzyme a2 synthetase 1 (apo form)
|
|
|
| 44 | c4qjiB_ |
|
not modelled |
95.0 |
24 |
PDB header:ligase
Chain: B: PDB Molecule:phosphopantothenate--cysteine ligase;
PDBTitle: crystal structure of the c-terminal ctp-binding domain of a2 phosphopantothenoylcysteine decarboxylase/phosphopantothenate-3 cysteine ligase with bound ctp from mycobacterium smegmatis
|
|
|
| 45 | c3x2fA_ |
|
not modelled |
94.2 |
13 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: a thermophilic s-adenosylhomocysteine hydrolase
|
|
|
| 46 | c5v96A_ |
|
not modelled |
94.2 |
15 |
PDB header:hydrolase
Chain: A: PDB Molecule:s-adenosyl-l-homocysteine hydrolase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from2 naegleria fowleri with bound nad and adenosine
|
|
|
| 47 | c1d4fD_ |
|
not modelled |
93.8 |
16 |
PDB header:hydrolase
Chain: D: PDB Molecule:s-adenosylhomocysteine hydrolase;
PDBTitle: crystal structure of recombinant rat-liver d244e mutant s-2 adenosylhomocysteine hydrolase
|
|
|
| 48 | c4r3uD_ |
|
not modelled |
93.6 |
17 |
PDB header:isomerase
Chain: D: PDB Molecule:2-hydroxyisobutyryl-coa mutase small subunit;
PDBTitle: crystal structure of 2-hydroxyisobutyryl-coa mutase
|
|
|
| 49 | c3n58D_ |
|
not modelled |
93.4 |
18 |
PDB header:hydrolase
Chain: D: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of s-adenosyl-l-homocysteine hydrolase from brucella2 melitensis in ternary complex with nad and adenosine, orthorhombic3 form
|
|
|
| 50 | d1u7za_ |
|
not modelled |
93.3 |
17 |
Fold:Ribokinase-like
Superfamily:CoaB-like
Family:CoaB-like |
|
|
| 51 | d1m1nb_ |
|
not modelled |
93.1 |
10 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 52 | c2rirA_ |
|
not modelled |
93.1 |
14 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dipicolinate synthase, a chain;
PDBTitle: crystal structure of dipicolinate synthase, a chain, from bacillus2 subtilis
|
|
|
| 53 | c3k9cA_ |
|
not modelled |
93.0 |
11 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family protein;
PDBTitle: crystal structure of laci transcriptional regulator from rhodococcus2 species.
|
|
|
| 54 | c3pdiB_ |
|
not modelled |
92.9 |
14 |
PDB header:protein binding
Chain: B: PDB Molecule:nitrogenase mofe cofactor biosynthesis protein nifn;
PDBTitle: precursor bound nifen
|
|
|
| 55 | c4njmA_ |
|
not modelled |
92.4 |
10 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-3-phosphoglycerate dehydrogenase, putative;
PDBTitle: crystal structure of phosphoglycerate bound 3-phosphoglycerate2 dehydrogenase in entamoeba histolytica
|
|
|
| 56 | d7reqa2 |
|
not modelled |
92.1 |
19 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 57 | d1ccwa_ |
|
not modelled |
91.9 |
13 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 58 | c3brqA_ |
|
not modelled |
91.8 |
10 |
PDB header:transcription regulator
Chain: A: PDB Molecule:hth-type transcriptional regulator ascg;
PDBTitle: crystal structure of the escherichia coli transcriptional repressor2 ascg
|
|
|
| 59 | c3dhyC_ |
|
not modelled |
91.5 |
19 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structures of mycobacterium tuberculosis s-adenosyl-l-2 homocysteine hydrolase in ternary complex with substrate and3 inhibitors
|
|
|
| 60 | c4rk1F_ |
|
not modelled |
91.2 |
9 |
PDB header:transcription regulator
Chain: F: PDB Molecule:ribose transcriptional regulator;
PDBTitle: crystal structure of laci family transcriptional regulator from2 enterococcus faecium, target efi-512930, with bound ribose
|
|
|
| 61 | c3clkB_ |
|
not modelled |
91.1 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcription regulator;
PDBTitle: crystal structure of a transcription regulator from lactobacillus2 plantarum
|
|
|
| 62 | c5intB_ |
|
not modelled |
90.8 |
21 |
PDB header:ligase
Chain: B: PDB Molecule:phosphopantothenate--cysteine ligase;
PDBTitle: crystal structure of the c-terminal domain of coenzyme a biosynthesis2 bifunctional protein coabc
|
|
|
| 63 | c3d8uA_ |
|
not modelled |
90.4 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:purr transcriptional regulator;
PDBTitle: the crystal structure of a purr family transcriptional regulator from2 vibrio parahaemolyticus rimd 2210633
|
|
|
| 64 | d1xu9a_ |
|
not modelled |
90.1 |
24 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 65 | c3qk7C_ |
|
not modelled |
89.8 |
8 |
PDB header:transcription regulator
Chain: C: PDB Molecule:transcriptional regulators;
PDBTitle: crystal structure of putative transcriptional regulator from yersinia2 pestis biovar microtus str. 91001
|
|
|
| 66 | d1zema1 |
|
not modelled |
89.2 |
14 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 67 | c5bt9B_ |
|
not modelled |
88.9 |
23 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of folm alternative dihydrofolate reductase 1 from2 brucella canis complexed with nadp
|
|
|
| 68 | c5tchG_ |
|
not modelled |
88.9 |
16 |
PDB header:lyase
Chain: G: PDB Molecule:tryptophan synthase alpha chain;
PDBTitle: crystal structure of tryptophan synthase from m. tuberculosis -2 ligand-free form, trpa-g66v mutant
|
|
|
| 69 | c3gvpB_ |
|
not modelled |
88.9 |
17 |
PDB header:hydrolase
Chain: B: PDB Molecule:adenosylhomocysteinase 3;
PDBTitle: human sahh-like domain of human adenosylhomocysteinase 3
|
|
|
| 70 | c3oneA_ |
|
not modelled |
88.9 |
16 |
PDB header:hydrolase/hydrolase substrate
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of lupinus luteus s-adenosyl-l-homocysteine2 hydrolase in complex with adenine
|
|
|
| 71 | c4n7bA_ |
|
not modelled |
88.8 |
9 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:lytb;
PDBTitle: structure of the e-1-hydroxy-2-methyl-but-2-enyl-4-diphosphate2 reductase from plasmodium falciparum
|
|
|
| 72 | c3q2oB_ |
|
not modelled |
88.2 |
9 |
PDB header:lyase
Chain: B: PDB Molecule:phosphoribosylaminoimidazole carboxylase, atpase subunit;
PDBTitle: crystal structure of purk: n5-carboxyaminoimidazole ribonucleotide2 synthetase
|
|
|
| 73 | d1fmfa_ |
|
not modelled |
88.1 |
12 |
Fold:Flavodoxin-like
Superfamily:Cobalamin (vitamin B12)-binding domain
Family:Cobalamin (vitamin B12)-binding domain |
|
|
| 74 | d1ys7a2 |
|
not modelled |
88.0 |
21 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 75 | c3ek2D_ |
|
not modelled |
87.6 |
11 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:enoyl-(acyl-carrier-protein) reductase (nadh);
PDBTitle: crystal structure of eonyl-(acyl carrier protein) reductase from2 burkholderia pseudomallei 1719b
|
|
|
| 76 | c4nbtA_ |
|
not modelled |
87.5 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-[acyl-carrier-protein] reductase;
PDBTitle: crystal structure of fabg from acholeplasma laidlawii
|
|
|
| 77 | c3h5oB_ |
|
not modelled |
87.3 |
10 |
PDB header:transcription regulator
Chain: B: PDB Molecule:transcriptional regulator gntr;
PDBTitle: the crystal structure of transcription regulator gntr from2 chromobacterium violaceum
|
|
|
| 78 | d2o8ra3 |
|
not modelled |
87.0 |
18 |
Fold:Phospholipase D/nuclease
Superfamily:Phospholipase D/nuclease
Family:Polyphosphate kinase C-terminal domain |
|
|
| 79 | c5v1tA_ |
|
not modelled |
86.7 |
15 |
PDB header:metal binding protein
Chain: A: PDB Molecule:radical sam;
PDBTitle: crystal structure of streptococcus suis suib bound to precursor2 peptide suia
|
|
|
| 80 | c3ke8A_ |
|
not modelled |
86.7 |
17 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:4-hydroxy-3-methylbut-2-enyl diphosphate reductase;
PDBTitle: crystal structure of isph:hmbpp-complex
|
|
|
| 81 | c3ksmA_ |
|
not modelled |
86.4 |
10 |
PDB header:transport protein
Chain: A: PDB Molecule:abc-type sugar transport system, periplasmic component;
PDBTitle: crystal structure of abc-type sugar transport system, periplasmic2 component from hahella chejuensis
|
|
|
| 82 | c4zgwB_ |
|
not modelled |
86.2 |
24 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:short-chain dehydrogenase/reductase;
PDBTitle: short-chain dehydrogenase/reductase from serratia marcescens bcrc2 10948
|
|
|
| 83 | c4weoD_ |
|
not modelled |
86.2 |
18 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:putative acetoin(diacetyl) reductase;
PDBTitle: crystal structure of a putative acetoin(diacetyl) reductase2 burkholderia cenocepacia
|
|
|
| 84 | c6hyhA_ |
|
not modelled |
85.7 |
10 |
PDB header:sugar binding protein
Chain: A: PDB Molecule:periplasmic binding protein/laci transcriptional regulator;
PDBTitle: crystal structure of msmeg_1712 from mycobacterium smegmatis in2 complex with beta-d-fucofuranose
|
|
|
| 85 | c5hm8C_ |
|
not modelled |
85.5 |
16 |
PDB header:hydrolase
Chain: C: PDB Molecule:adenosylhomocysteinase;
PDBTitle: 2.85 angstrom crystal structure of s-adenosylhomocysteinase from2 cryptosporidium parvum in complex with adenosine and nad.
|
|
|
| 86 | c1v8bA_ |
|
not modelled |
85.5 |
14 |
PDB header:hydrolase
Chain: A: PDB Molecule:adenosylhomocysteinase;
PDBTitle: crystal structure of a hydrolase
|
|
|
| 87 | c4g2nA_ |
|
not modelled |
85.4 |
13 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:d-isomer specific 2-hydroxyacid dehydrogenase, nad-binding;
PDBTitle: crystal structure of putative d-isomer specific 2-hydroxyacid2 dehydrogenase, nad-binding from polaromonas sp. js6 66
|
|
|
| 88 | c4imrA_ |
|
not modelled |
85.3 |
29 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-oxoacyl-(acyl-carrier-protein) reductase;
PDBTitle: crystal structure of 3-oxoacyl (acyl-carrier-protein) reductase2 (target efi-506442) from agrobacterium tumefaciens c58 with nadp3 bound
|
|
|
| 89 | c5unlA_ |
|
not modelled |
85.2 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:3-ketoacyl-acp reductase;
PDBTitle: crystal structure of a d-beta-hydroxybutyrate dehydrogenase from2 burkholderia multivorans
|
|
|
| 90 | c3k4hA_ |
|
not modelled |
85.0 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:putative transcriptional regulator;
PDBTitle: crystal structure of putative transcriptional regulator laci from2 bacillus cereus subsp. cytotoxis nvh 391-98
|
|
|
| 91 | c3g85A_ |
|
not modelled |
85.0 |
9 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator (laci family);
PDBTitle: crystal structure of laci family transcription regulator from2 clostridium acetobutylicum
|
|
|
| 92 | c2qq5A_ |
|
not modelled |
84.7 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:dehydrogenase/reductase sdr family member 1;
PDBTitle: crystal structure of human sdr family member 1
|
|
|
| 93 | c4yt2A_ |
|
not modelled |
84.7 |
21 |
PDB header:metal binding protein
Chain: A: PDB Molecule:h(2)-forming methylenetetrahydromethanopterin
PDBTitle: hmd ii from methanocaldococcus jannaschii
|
|
|
| 94 | d1mvoa_ |
|
not modelled |
84.7 |
9 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 95 | c5jydA_ |
|
not modelled |
84.4 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase;
PDBTitle: crystal structure of a putative short chain dehydrogenase from2 burkholderia cenocepacia
|
|
|
| 96 | c6huxA_ |
|
not modelled |
84.4 |
21 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:h(2)-forming methylenetetrahydromethanopterin
PDBTitle: hmdii from methanocaldococcus jannaschii reconstitued with fe-2 guanylylpyridinol (fegp) cofactor and co-crystallized with methenyl-3 tetrahydromethanopterin at 2.5 a resolution
|
|
|
| 97 | d1ja9a_ |
|
not modelled |
84.0 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 98 | c4b79B_ |
|
not modelled |
83.6 |
19 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:probable short-chain dehydrogenase;
PDBTitle: the aeropath project and pseudomonas aeruginosa high-throughput2 crystallographic studies for assessment of potential targets in3 early stage drug discovery.
|
|
|
| 99 | c3c3kA_ |
|
not modelled |
83.6 |
11 |
PDB header:isomerase
Chain: A: PDB Molecule:alanine racemase;
PDBTitle: crystal structure of an uncharacterized protein from actinobacillus2 succinogenes
|
|
|
| 100 | d1eo1a_ |
|
not modelled |
83.5 |
22 |
Fold:Ribonuclease H-like motif
Superfamily:Nitrogenase accessory factor-like
Family:MTH1175-like |
|
|
| 101 | c2o8rA_ |
|
not modelled |
83.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:polyphosphate kinase;
PDBTitle: crystal structure of polyphosphate kinase from porphyromonas2 gingivalis
|
|
|
| 102 | c5k9zB_ |
|
not modelled |
83.0 |
18 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:putative short-chain dehydrogenase/reductase;
PDBTitle: crystal structure of putative short-chain dehydrogenase/reductase from2 burkholderia xenovorans lb400
|
|
|
| 103 | c5fydB_ |
|
not modelled |
82.9 |
26 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:oxidoreductase, short chain dehydrogenase/reductase
PDBTitle: structural and biochemical insights into 7beta-2 hydroxysteroid dehydrogenase stereoselectivity
|
|
|
| 104 | c3crnA_ |
|
not modelled |
82.9 |
18 |
PDB header:signaling protein
Chain: A: PDB Molecule:response regulator receiver domain protein, chey-like;
PDBTitle: crystal structure of response regulator receiver domain protein (chey-2 like) from methanospirillum hungatei jf-1
|
|
|
| 105 | d1geea_ |
|
not modelled |
82.7 |
21 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 106 | c3rfqC_ |
|
not modelled |
82.7 |
22 |
PDB header:biosynthetic protein
Chain: C: PDB Molecule:pterin-4-alpha-carbinolamine dehydratase moab2;
PDBTitle: crystal structure of pterin-4-alpha-carbinolamine dehydratase moab22 from mycobacterium marinum
|
|
|
| 107 | c2mswA_ |
|
not modelled |
82.6 |
9 |
PDB header:transferase
Chain: A: PDB Molecule:response regulator/sensor histidine kinase;
PDBTitle: ligand-induced folding of a receiver domain
|
|
|
| 108 | d2d1ya1 |
|
not modelled |
82.5 |
22 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 109 | d1qh8b_ |
|
not modelled |
82.4 |
11 |
Fold:Chelatase-like
Superfamily:"Helical backbone" metal receptor
Family:Nitrogenase iron-molybdenum protein |
|
|
| 110 | d1krwa_ |
|
not modelled |
82.4 |
13 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 111 | c5th5C_ |
|
not modelled |
82.3 |
8 |
PDB header:lyase
Chain: C: PDB Molecule:7-carboxy-7-deazaguanine synthase;
PDBTitle: crystal structure of quee from bacillus subtilis with 6-carboxypterin-2 5'-deoxyadenosyl ester bound
|
|
|
| 112 | c4h2dB_ |
|
not modelled |
82.2 |
10 |
PDB header:oxidoreductase
Chain: B: PDB Molecule:nadph-dependent diflavin oxidoreductase 1;
PDBTitle: crystal structure of ndor1
|
|
|
| 113 | d2ayxa1 |
|
not modelled |
82.0 |
15 |
Fold:Flavodoxin-like
Superfamily:CheY-like
Family:CheY-related |
|
|
| 114 | c3fbtB_ |
|
not modelled |
81.8 |
11 |
PDB header:oxidoreductase, lyase
Chain: B: PDB Molecule:chorismate mutase and shikimate 5-dehydrogenase fusion
PDBTitle: crystal structure of a chorismate mutase/shikimate 5-dehydrogenase2 fusion protein from clostridium acetobutylicum
|
|
|
| 115 | d1hxha_ |
|
not modelled |
81.8 |
23 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 116 | d1fmca_ |
|
not modelled |
81.7 |
17 |
Fold:NAD(P)-binding Rossmann-fold domains
Superfamily:NAD(P)-binding Rossmann-fold domains
Family:Tyrosine-dependent oxidoreductases |
|
|
| 117 | c6c8vA_ |
|
not modelled |
81.7 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:coenzyme pqq synthesis protein e;
PDBTitle: x-ray structure of pqqe from methylobacterium extorquens
|
|
|
| 118 | c4fn4A_ |
|
not modelled |
81.6 |
15 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:short chain dehydrogenase;
PDBTitle: short-chain nad(h)-dependent dehydrogenase/reductase from sulfolobus2 acidocaldarius
|
|
|
| 119 | c2hrzA_ |
|
not modelled |
81.5 |
14 |
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:nucleoside-diphosphate-sugar epimerase;
PDBTitle: the crystal structure of the nucleoside-diphosphate-sugar epimerase2 from agrobacterium tumefaciens
|
|
|
| 120 | c3gv0A_ |
|
not modelled |
81.3 |
13 |
PDB header:transcription regulator
Chain: A: PDB Molecule:transcriptional regulator, laci family;
PDBTitle: crystal structure of laci family transcription regulator from2 agrobacterium tumefaciens
|
|
|