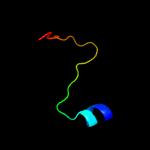
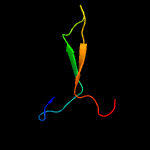
1 d3ci0k1
20.2
67
Fold: SAM domain-likeSuperfamily: GspK insert domain-likeFamily: GspK insert domain-like
2 c2lorA_
20.2
27
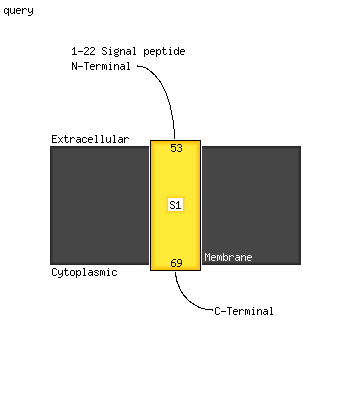
PDB header: membrane proteinChain: A: PDB Molecule: transmembrane protein 141;PDBTitle: backbone structure of human membrane protein tmem141
3 c5zhhB_
14.5
35
PDB header: hydrolaseChain: B: PDB Molecule: inositol-1-monophosphatase;PDBTitle: structure of inositol monophosphatase from anabaena (nostoc) sp. pcc2 7120
4 d1isua_
11.8
58
Fold: HIPIP (high potential iron protein)Superfamily: HIPIP (high potential iron protein)Family: HIPIP (high potential iron protein)
5 c5z62N_
9.5
38
PDB header: electron transportChain: N: PDB Molecule: cytochrome c oxidase subunit ndufa4;PDBTitle: structure of human cytochrome c oxidase
6 d1vdwa_
8.5
30
Fold: Carbohydrate phosphataseSuperfamily: Carbohydrate phosphataseFamily: Inositol monophosphatase/fructose-1,6-bisphosphatase-like
7 c2pcrA_
8.2
35
PDB header: hydrolaseChain: A: PDB Molecule: inositol-1-monophosphatase;PDBTitle: crystal structure of myo-inositol-1(or 4)-monophosphatase (aq_1983)2 from aquifex aeolicus vf5
8 d1f5ma_
8.2
16
Fold: Profilin-likeSuperfamily: GAF domain-likeFamily: GAF domain
9 d2pkha1
8.0
11
Fold: Chorismate lyase-likeSuperfamily: Chorismate lyase-likeFamily: UTRA domain
10 c2gk2B_
7.9
41
PDB header: cell adhesionChain: B: PDB Molecule: carcinoembryonic antigen-related cell adhesion molecule 1;PDBTitle: crystal structure of the n terminal domain of human ceacam1
11 c5eq9A_
7.4
25
PDB header: biosynthetic proteinChain: A: PDB Molecule: inositol monophosphatase;PDBTitle: crystal structure of medicago truncatula histidinol-phosphate2 phosphatase (mthpp) in complex with l-histidinol phosphate and mg2+
12 c4lduA_
7.2
60
PDB header: dna binding proteinChain: A: PDB Molecule: auxin response factor 5;PDBTitle: crystal structure of the dna binding domain of arabidopsis thaliana2 auxin response factor 5
13 c3s9vD_
7.1
40
PDB header: lyase, isomeraseChain: D: PDB Molecule: abietadiene synthase, chloroplastic;PDBTitle: abietadiene synthase from abies grandis
14 c4ldxB_
7.0
56
PDB header: transcription/dnaChain: B: PDB Molecule: auxin response factor 1;PDBTitle: crystal structure of the dna binding domain of arabidopsis thaliana2 auxin response factor 1 (arf1) in complex with protomor-like sequence3 er7
15 c2dksA_
6.9
31
PDB header: cell adhesionChain: A: PDB Molecule: carcinoembryonic antigen-related cell adhesionPDBTitle: solution structure of the first ig-like domain of human2 carcinoembryonic antigen related cell adhesion molecule 8
16 d1rk8c_
6.7
57
Fold: WW domain-likeSuperfamily: Pym (Within the bgcn gene intron protein, WIBG), N-terminal domainFamily: Pym (Within the bgcn gene intron protein, WIBG), N-terminal domain
17 c1rk8C_
6.7
57
PDB header: translationChain: C: PDB Molecule: within the bgcn gene intron protein;PDBTitle: structure of the cytosolic protein pym bound to the mago-2 y14 core of the exon junction complex
18 c2jijA_
6.6
25
PDB header: hydrolaseChain: A: PDB Molecule: prolyl-4 hydroxylase;PDBTitle: crystal structure of the apo form of chlamydomonas2 reinhardtii prolyl-4 hydroxylase type i
19 c3pybB_
5.9
40
PDB header: isomeraseChain: B: PDB Molecule: ent-copalyl diphosphate synthase, chloroplastic;PDBTitle: crystal structure of ent-copalyl diphosphate synthase from arabidopsis2 thaliana in complex with 13-aza-13,14-dihydrocopalyl diphosphate
20 c4qycA_
5.8
41
PDB header: cell adhesionChain: A: PDB Molecule: carcinoembryonic antigen-related cell adhesion molecule 1,PDBTitle: crystal structure of the chimeric protein human ceacam1: human tim32 membrane distal amino terminal (n)-domain
21 d1l6za1
not modelled
5.7
33
Fold: Immunoglobulin-like beta-sandwichSuperfamily: ImmunoglobulinFamily: V set domains (antibody variable domain-like)
22 c2czhB_
not modelled
5.7
20
PDB header: hydrolaseChain: B: PDB Molecule: inositol monophosphatase 2;PDBTitle: crystal structure of human myo-inositol monophosphatase 22 (impa2) with phosphate ion (orthorhombic form)
23 c3u6gB_
not modelled
5.5
26
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein duf4425;PDBTitle: crystal structure of a domain of unknown function, duf4425 (bvu_3708)2 from bacteroides vulgatus atcc 8482 at 1.35 a resolution
24 c3p5rB_
not modelled
5.4
44
PDB header: lyaseChain: B: PDB Molecule: taxadiene synthase;PDBTitle: crystal structure of taxadiene synthase from pacific yew (taxus2 brevifolia) in complex with mg2+ and 2-fluorogeranylgeranyl3 diphosphate
25 c5bp8A_
not modelled
5.3
42
PDB header: isomeraseChain: A: PDB Molecule: ent-copalyl diphosphate synthase;PDBTitle: ent-copalyl diphosphate synthase from streptomyces platensis
26 d1c3pa_
not modelled
5.2
50
Fold: Arginase/deacetylaseSuperfamily: Arginase/deacetylaseFamily: Histone deacetylase, HDAC
27 d1mpga2
not modelled
5.1
21
Fold: TBP-likeSuperfamily: TATA-box binding protein-likeFamily: DNA repair glycosylase, N-terminal domain