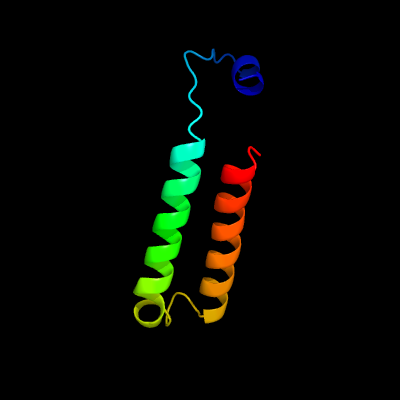
| 1 | d2o57a1
|
|
 |
98.8 |
30 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
|
|
|
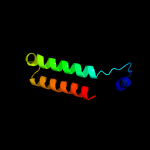
| 2 | c5gm2E_
|
|
 |
97.3 |
21 |
PDB header:transferase
Chain: E: PDB Molecule:o-methylransferase;
PDBTitle: crystal structure of methyltransferase tled complexed with sah and2 teleocidin a1
|
|
|
|
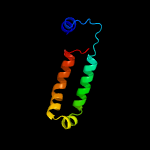
| 3 | c5wp5A_
|
|
 |
97.0 |
19 |
PDB header:transferase
Chain: A: PDB Molecule:phosphomethylethanolamine n-methyltransferase 2;
PDBTitle: arabidopsis thaliana phosphoethanolamine n-methyltransferase 22 (atpmt2) in complex with sah
|
|
|
|
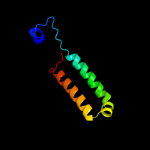
| 4 | c4iv0B_
|
|
 |
96.9 |
11 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoethanolamine n-methyltransferase, putative;
PDBTitle: crystal structure of n-methyl transferase from plasmodium vivax2 complexed with s-adenosyl methionine and phosphate
|
|
|
|
| 5 | c2fk8A_
|
|
 |
96.8 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:methoxy mycolic acid synthase 4;
PDBTitle: crystal structure of hma (mmaa4) from mycobacterium tuberculosis2 complexed with s-adenosylmethionine
|
|
|
|
| 6 | d1tpya_
|
|
 |
96.2 |
17 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
|
|
|
| 7 | d1kpga_
|
|
 |
95.5 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
|
|
|
| 8 | c3busB_
|
|
 |
95.4 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:methyltransferase;
PDBTitle: crystal structure of rebm
|
|
|
|
| 9 | d2fk8a1
|
|
 |
95.3 |
14 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
|
|
|
| 10 | d1l1ea_
|
|
 |
94.8 |
21 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
|
|
|
| 11 | c4krhB_
|
|
 |
93.5 |
18 |
PDB header:transferase
Chain: B: PDB Molecule:phosphoethanolamine n-methyltransferase 2;
PDBTitle: semet haemonchus contortus phosphoethanolamine n-methyltransferase 22 in complex with s-adenosyl-l-methionine
|
|
|
|
| 12 | d1kpia_
|
|
 |
93.1 |
16 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Mycolic acid cyclopropane synthase |
|
|
|
| 13 | c4ineB_
|
|
 |
93.0 |
14 |
PDB header:transferase
Chain: B: PDB Molecule:protein pmt-2;
PDBTitle: crystal structure of n-methyl transferase (pmt-2) from caenorhabditis2 elegant complexed with s-adenosyl homocysteine and3 phosphoethanolamine
|
|
|
|
| 14 | c5z9oA_
|
|
 |
92.7 |
18 |
PDB header:transferase
Chain: A: PDB Molecule:cyclopropane-fatty-acyl-phospholipid synthase;
PDBTitle: the crystal structure of cyclopropane-fatty-acyl-phospholipid synthase2 from lactobacillus acidophilus
|
|
|
|
| 15 | c3ujcA_
|
|
 |
92.3 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:phosphoethanolamine n-methyltransferase;
PDBTitle: phosphoethanolamine methyltransferase mutant (h132a) from plasmodium2 falciparum in complex with phosphocholine
|
|
|
|
| 16 | c4pneA_
|
|
 |
89.4 |
24 |
PDB header:biosynthetic protein
Chain: A: PDB Molecule:methyltransferase-like protein;
PDBTitle: crystal structure of the [4+2]-cyclase spnf
|
|
|
|
| 17 | c6bqcA_
|
|
 |
78.5 |
14 |
PDB header:transferase
Chain: A: PDB Molecule:cyclopropane-fatty-acyl-phospholipid synthase;
PDBTitle: cyclopropane fatty acid synthase from e. coli
|
|
|
|
| 18 | d1nkva_
|
|
 |
75.7 |
19 |
Fold:S-adenosyl-L-methionine-dependent methyltransferases
Superfamily:S-adenosyl-L-methionine-dependent methyltransferases
Family:Hypothetical Protein YjhP |
|
|
|
| 19 | d1q08a_
|
|
 |
43.4 |
21 |
Fold:Putative DNA-binding domain
Superfamily:Putative DNA-binding domain
Family:DNA-binding N-terminal domain of transcription activators |
|
|
|
| 20 | c2rq5A_
|
|
 |
34.5 |
17 |
PDB header:transcription
Chain: A: PDB Molecule:protein jumonji;
PDBTitle: solution structure of the at-rich interaction domain (arid)2 of jumonji/jarid2
|
|
|
|
| 21 | c6nuwG_ |
|
not modelled |
33.0 |
13 |
PDB header:cell cycle
Chain: G: PDB Molecule:inner kinetochore subunit nkp1;
PDBTitle: yeast ctf19 complex
|
|
|
| 22 | c1ye9E_ |
|
not modelled |
32.1 |
17 |
PDB header:oxidoreductase
Chain: E: PDB Molecule:catalase hpii;
PDBTitle: crystal structure of proteolytically truncated catalase2 hpii from e. coli
|
|
|
| 23 | c2jrzA_ |
|
not modelled |
29.3 |
18 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:histone demethylase jarid1c;
PDBTitle: solution structure of the bright/arid domain from the human2 jarid1c protein.
|
|
|
| 24 | c4p47A_ |
|
not modelled |
28.3 |
15 |
PDB header:solute-binding protein
Chain: A: PDB Molecule:trap dicarboxylate transporter, dctp subunit;
PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 ochrobactrum anthropi (oant_4429), target efi-510151, c-termius bound3 in ligand binding pocket
|
|
|
| 25 | c4qoqC_ |
|
not modelled |
28.3 |
15 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:catalase;
PDBTitle: structure of bacillus pumilus catalase with guaiacol bound
|
|
|
| 26 | d1e93a_ |
|
not modelled |
24.1 |
17 |
Fold:Heme-dependent catalase-like
Superfamily:Heme-dependent catalase-like
Family:Heme-dependent catalases |
|
|
| 27 | d1ig8a1 |
|
not modelled |
22.9 |
13 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
|
|
| 28 | d1yoza1 |
|
not modelled |
20.7 |
31 |
Fold:AF0941-like
Superfamily:AF0941-like
Family:AF0941-like |
|
|
| 29 | c1f4jC_ |
|
not modelled |
20.0 |
10 |
PDB header:oxidoreductase
Chain: C: PDB Molecule:catalase;
PDBTitle: structure of tetragonal crystals of human erythrocyte catalase
|
|
|
| 30 | c4cabA_ |
|
not modelled |
19.4 |
23 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:catalase;
PDBTitle: the refined structure of catalase dr1998 from deinococcus radiodurans2 at 2.6 a resolution
|
|
|
| 31 | d1m7sa_ |
|
not modelled |
19.0 |
17 |
Fold:Heme-dependent catalase-like
Superfamily:Heme-dependent catalase-like
Family:Heme-dependent catalases |
|
|
| 32 | c3e4cB_ |
|
not modelled |
17.5 |
29 |
PDB header:hydrolase
Chain: B: PDB Molecule:caspase-1;
PDBTitle: procaspase-1 zymogen domain crystal strucutre
|
|
|
| 33 | c3h11A_ |
|
not modelled |
17.0 |
30 |
PDB header:apoptosis
Chain: A: PDB Molecule:casp8 and fadd-like apoptosis regulator;
PDBTitle: zymogen caspase-8:c-flipl protease domain complex
|
|
|
| 34 | d2io8a1 |
|
not modelled |
16.3 |
35 |
Fold:PreATP-grasp domain
Superfamily:PreATP-grasp domain
Family:Glutathionylspermidine synthase substrate-binding domain-like |
|
|
| 35 | d1dgfa_ |
|
not modelled |
16.2 |
10 |
Fold:Heme-dependent catalase-like
Superfamily:Heme-dependent catalase-like
Family:Heme-dependent catalases |
|
|
| 36 | c2xq1M_ |
|
not modelled |
15.7 |
24 |
PDB header:oxidoreductase
Chain: M: PDB Molecule:peroxisomal catalase;
PDBTitle: crystal structure of peroxisomal catalase from the yeast hansenula2 polymorpha
|
|
|
| 37 | d2j78a1 |
|
not modelled |
15.7 |
26 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
|
|
| 38 | d1uwsa_ |
|
not modelled |
15.4 |
29 |
Fold:TIM beta/alpha-barrel
Superfamily:(Trans)glycosidases
Family:Family 1 of glycosyl hydrolase |
|
|
| 39 | c6g21B_ |
|
not modelled |
15.3 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:probable feruloyl esterase b-2;
PDBTitle: crystal structure of an esterase from aspergillus oryzae
|
|
|
| 40 | d1fp1d1 |
|
not modelled |
15.1 |
18 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:"Winged helix" DNA-binding domain
Family:Plant O-methyltransferase, N-terminal domain |
|
|
| 41 | c6c4rA_ |
|
not modelled |
14.7 |
24 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:staphylopine dehydrogenase;
PDBTitle: staphylopine dehydrogenase (saodh) - apo
|
|
|
| 42 | c1ig8A_ |
|
not modelled |
14.4 |
13 |
PDB header:transferase
Chain: A: PDB Molecule:hexokinase pii;
PDBTitle: crystal structure of yeast hexokinase pii with the correct2 amino acid sequence
|
|
|
| 43 | c2wsiA_ |
|
not modelled |
14.4 |
15 |
PDB header:transferase
Chain: A: PDB Molecule:fad synthetase;
PDBTitle: crystal structure of yeast fad synthetase (fad1) in complex2 with fad
|
|
|
| 44 | c4wwrB_ |
|
not modelled |
14.4 |
21 |
PDB header:transport protein
Chain: B: PDB Molecule:ubiquitin-like protein 4a;
PDBTitle: crystal structure of bag6-ubl4a dimerization domain
|
|
|
| 45 | c1cp3B_ |
|
not modelled |
14.2 |
38 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:apopain;
PDBTitle: crystal structure of the complex of apopain with the tetrapeptide2 inhibitor ace-dvad-fmc
|
|
|
| 46 | c2fp3A_ |
|
not modelled |
14.1 |
14 |
PDB header:hydrolysis/apoptosis
Chain: A: PDB Molecule:caspase nc;
PDBTitle: crystal structure of the drosophila initiator caspase dronc
|
|
|
| 47 | d1gwea_ |
|
not modelled |
14.0 |
17 |
Fold:Heme-dependent catalase-like
Superfamily:Heme-dependent catalase-like
Family:Heme-dependent catalases |
|
|
| 48 | d1c20a_ |
|
not modelled |
14.0 |
15 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:ARID-like
Family:ARID domain |
|
|
| 49 | c2yqeA_ |
|
not modelled |
13.6 |
28 |
PDB header:dna binding protein
Chain: A: PDB Molecule:jumonji/arid domain-containing protein 1d;
PDBTitle: solution structure of the arid domain of jarid1d protein
|
|
|
| 50 | c4iyrB_ |
|
not modelled |
13.6 |
35 |
PDB header:hydrolase
Chain: B: PDB Molecule:caspase-6;
PDBTitle: crystal structure of full-length caspase-6 zymogen
|
|
|
| 51 | c1kmcB_ |
|
not modelled |
13.4 |
24 |
PDB header:apoptosis/hydrolase
Chain: B: PDB Molecule:caspase-7;
PDBTitle: crystal structure of the caspase-7 / xiap-bir2 complex
|
|
|
| 52 | c6fatA_ |
|
not modelled |
13.2 |
20 |
PDB header:hydrolase
Chain: A: PDB Molecule:carboxylic ester hydrolase;
PDBTitle: the crystal structure of a feruloyl esterase c from fusarium2 oxysporum.
|
|
|
| 53 | d1q9ua_ |
|
not modelled |
13.1 |
29 |
Fold:TBP-like
Superfamily:TT1751-like
Family:TT1751-like |
|
|
| 54 | c4wwrD_ |
|
not modelled |
13.1 |
21 |
PDB header:transport protein
Chain: D: PDB Molecule:ubiquitin-like protein 4a;
PDBTitle: crystal structure of bag6-ubl4a dimerization domain
|
|
|
| 55 | c1ysmA_ |
|
not modelled |
12.7 |
23 |
PDB header:metal binding protein
Chain: A: PDB Molecule:calcyclin-binding protein;
PDBTitle: nmr structure of n-terminal domain (residues 1-77) of siah-2 interacting protein.
|
|
|
| 56 | d1ysma1 |
|
not modelled |
12.7 |
23 |
Fold:Long alpha-hairpin
Superfamily:Calcyclin-binding protein-like
Family:Siah interacting protein N terminal domain-like |
|
|
| 57 | d1si8a_ |
|
not modelled |
12.6 |
21 |
Fold:Heme-dependent catalase-like
Superfamily:Heme-dependent catalase-like
Family:Heme-dependent catalases |
|
|
| 58 | c1nmqB_ |
|
not modelled |
12.6 |
38 |
PDB header:apoptosis, hydrolase
Chain: B: PDB Molecule:caspase-3;
PDBTitle: extendend tethering: in situ assembly of inhibitors
|
|
|
| 59 | d2d5ua1 |
|
not modelled |
12.5 |
16 |
Fold:PUG domain-like
Superfamily:PUG domain-like
Family:PUG domain |
|
|
| 60 | c4pakA_ |
|
not modelled |
12.5 |
22 |
PDB header:solute-binding protein
Chain: A: PDB Molecule:trap dicarboxylate transporter, dctp subunit;
PDBTitle: crystal structure of a trap periplasmic solute binding protein from2 verminephrobacter eiseniae ef01-2 (veis_3954, target efi-510324) a3 nephridial symbiont of the earthworm eisenia foetida, bound to (r)-4 pantoic acid
|
|
|
| 61 | d1j3ma_ |
|
not modelled |
12.5 |
24 |
Fold:TBP-like
Superfamily:TT1751-like
Family:TT1751-like |
|
|
| 62 | d4blca_ |
|
not modelled |
12.2 |
13 |
Fold:Heme-dependent catalase-like
Superfamily:Heme-dependent catalase-like
Family:Heme-dependent catalases |
|
|
| 63 | d1m72a_ |
|
not modelled |
12.2 |
24 |
Fold:Caspase-like
Superfamily:Caspase-like
Family:Caspase catalytic domain |
|
|
| 64 | d1ig6a_ |
|
not modelled |
12.1 |
19 |
Fold:DNA/RNA-binding 3-helical bundle
Superfamily:ARID-like
Family:ARID domain |
|
|
| 65 | c5jftB_ |
|
not modelled |
12.1 |
19 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:caspase 3, apoptosis-related cysteine protease a;
PDBTitle: zebra fish caspase-3
|
|
|
| 66 | c3wmtA_ |
|
not modelled |
12.0 |
18 |
PDB header:hydrolase
Chain: A: PDB Molecule:probable feruloyl esterase b-1;
PDBTitle: crystal structure of feruloyl esterase b from aspergillus oryzae
|
|
|
| 67 | c4e1rA_ |
|
not modelled |
11.5 |
39 |
PDB header:dna binding protein
Chain: A: PDB Molecule:protein lsr2;
PDBTitle: crystal structure of the dimerization domain of lsr2 from2 mycobacterium tuberculosis in the p 31 2 1 space group
|
|
|
| 68 | c4e1pA_ |
|
not modelled |
11.5 |
39 |
PDB header:dna binding protein
Chain: A: PDB Molecule:protein lsr2;
PDBTitle: crystal structure of the dimerization domain of lsr2 from2 mycobacterium tuberculosis in the p 1 21 1 space group
|
|
|
| 69 | c3zey8_ |
|
not modelled |
11.4 |
46 |
PDB header:ribosome
Chain: 8: PDB Molecule:ribosomal protein s29, putative;
PDBTitle: high-resolution cryo-electron microscopy structure of the trypanosoma2 brucei ribosome
|
|
|
| 70 | c3uoaB_ |
|
not modelled |
10.4 |
27 |
PDB header:hydrolase/hydrolase inhibitor
Chain: B: PDB Molecule:mucosa-associated lymphoid tissue lymphoma translocation
PDBTitle: crystal structure of the malt1 paracaspase (p21 form)
|
|
|
| 71 | c2nn3D_ |
|
not modelled |
10.4 |
24 |
PDB header:hydrolase
Chain: D: PDB Molecule:caspase-1;
PDBTitle: structure of pro-sf-caspase-1
|
|
|
| 72 | c4m9rB_ |
|
not modelled |
10.4 |
15 |
PDB header:hydrolase
Chain: B: PDB Molecule:cell death protein 3;
PDBTitle: crystal structure of ced-3
|
|
|
| 73 | c5zigD_ |
|
not modelled |
10.3 |
26 |
PDB header:isomerase
Chain: D: PDB Molecule:cellobiose 2-epimerase;
PDBTitle: the structure of cellobiose 2-epimerase from spirochaeta thermophila2 dsm 6192
|
|
|
| 74 | c4r27B_ |
|
not modelled |
10.3 |
26 |
PDB header:hydrolase
Chain: B: PDB Molecule:glycoside hydrolase;
PDBTitle: crystal structure of beta-glycosidase bgl167
|
|
|
| 75 | d1czan1 |
|
not modelled |
10.1 |
12 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
|
|
| 76 | c1kmiZ_ |
|
not modelled |
10.1 |
24 |
PDB header:signaling protein
Chain: Z: PDB Molecule:chemotaxis protein chez;
PDBTitle: crystal structure of an e.coli chemotaxis protein, chez
|
|
|
| 77 | c1cw5A_ |
|
not modelled |
10.0 |
19 |
PDB header:toxin
Chain: A: PDB Molecule:type iia bacteriocin carnobacteriocin b2;
PDBTitle: solution structure of carnobacteriocin b2
|
|
|
| 78 | c3fxtB_ |
|
not modelled |
9.8 |
5 |
PDB header:gene regulation
Chain: B: PDB Molecule:nucleoside diphosphate-linked moiety x motif 6;
PDBTitle: crystal structure of the n-terminal domain of human nudt6
|
|
|
| 79 | d1nw9b_ |
|
not modelled |
9.8 |
30 |
Fold:Caspase-like
Superfamily:Caspase-like
Family:Caspase catalytic domain |
|
|
| 80 | c3edqC_ |
|
not modelled |
9.7 |
38 |
PDB header:hydrolase/hydrolase inhibitor
Chain: C: PDB Molecule:caspase-3;
PDBTitle: crystal structure of caspase-3 with inhibitor ac-ldesd-cho
|
|
|
| 81 | c2lm1A_ |
|
not modelled |
9.6 |
19 |
PDB header:dna binding protein
Chain: A: PDB Molecule:lysine-specific demethylase lid;
PDBTitle: solution nmr structure of lysine-specific demethylase lid from2 drosophila melanogaster, northeast structural genomics consortium3 target fr824d
|
|
|
| 82 | c2fipA_ |
|
not modelled |
9.6 |
24 |
PDB header:transcription
Chain: A: PDB Molecule:late genes activator;
PDBTitle: phage phi29 transcription regulator p4
|
|
|
| 83 | d1f1ja_ |
|
not modelled |
9.5 |
24 |
Fold:Caspase-like
Superfamily:Caspase-like
Family:Caspase catalytic domain |
|
|
| 84 | c5vakA_ |
|
not modelled |
9.5 |
18 |
PDB header:signaling protein
Chain: A: PDB Molecule:beta-klotho;
PDBTitle: crystal structure of beta-klotho, domain 1
|
|
|
| 85 | c2v7yA_ |
|
not modelled |
9.5 |
19 |
PDB header:chaperone
Chain: A: PDB Molecule:chaperone protein dnak;
PDBTitle: crystal structure of the molecular chaperone dnak from2 geobacillus kaustophilus hta426 in post-atp hydrolysis3 state
|
|
|
| 86 | c3sipC_ |
|
not modelled |
9.4 |
24 |
PDB header:hydrolase/ligase/hydrolase
Chain: C: PDB Molecule:caspase;
PDBTitle: crystal structure of drice and diap1-bir1 complex
|
|
|
| 87 | c2funB_ |
|
not modelled |
9.3 |
19 |
PDB header:apoptosis/hydrolase
Chain: B: PDB Molecule:caspase-8;
PDBTitle: alternative p35-caspase-8 complex
|
|
|
| 88 | c6qgbA_ |
|
not modelled |
9.3 |
28 |
PDB header:hydrolase
Chain: A: PDB Molecule:mono(2-hydroxyethyl) terephthalate hydrolase;
PDBTitle: crystal structure of ideonella sakaiensis mhetase bound to benzoic2 acid
|
|
|
| 89 | c2j2mD_ |
|
not modelled |
9.0 |
17 |
PDB header:oxidoreductase
Chain: D: PDB Molecule:catalase;
PDBTitle: crystal structure analysis of catalase from exiguobacterium2 oxidotolerans
|
|
|
| 90 | d1bg3a1 |
|
not modelled |
8.9 |
12 |
Fold:Ribonuclease H-like motif
Superfamily:Actin-like ATPase domain
Family:Hexokinase |
|
|
| 91 | c2wdpA_ |
|
not modelled |
8.9 |
33 |
PDB header:hydrolase
Chain: A: PDB Molecule:caspase-6;
PDBTitle: crystal structure of ligand free human caspase-6
|
|
|
| 92 | c1i51A_ |
|
not modelled |
8.9 |
24 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:caspase-7 subunit p20;
PDBTitle: crystal structure of caspase-7 complexed with xiap
|
|
|
| 93 | c5u1dX_ |
|
not modelled |
8.8 |
28 |
PDB header:transport protein
Chain: X: PDB Molecule:tap transporter inhibitor icp47;
PDBTitle: cryo-em structure of the human tap atp-binding cassette transporter
|
|
|
| 94 | d1x1ia1 |
|
not modelled |
8.6 |
12 |
Fold:alpha/alpha toroid
Superfamily:Chondroitin AC/alginate lyase
Family:Hyaluronate lyase-like catalytic, N-terminal domain |
|
|
| 95 | c2ql5A_ |
|
not modelled |
8.5 |
24 |
PDB header:hydrolase/hydrolase inhibitor
Chain: A: PDB Molecule:caspase-7;
PDBTitle: crystal structure of caspase-7 with inhibitor ac-dmqd-cho
|
|
|
| 96 | c6axfG_ |
|
not modelled |
8.3 |
12 |
PDB header:protein binding
Chain: G: PDB Molecule:ras guanyl-releasing protein 2;
PDBTitle: structure of rasgrp2 in complex with rap1b
|
|
|
| 97 | d1qwla_ |
|
not modelled |
8.2 |
17 |
Fold:Heme-dependent catalase-like
Superfamily:Heme-dependent catalase-like
Family:Heme-dependent catalases |
|
|
| 98 | c3p45I_ |
|
not modelled |
8.1 |
33 |
PDB header:hydrolase
Chain: I: PDB Molecule:caspase-6;
PDBTitle: crystal structure of apo-caspase-6 at physiological ph
|
|
|
| 99 | c2jxjA_ |
|
not modelled |
7.9 |
22 |
PDB header:oxidoreductase
Chain: A: PDB Molecule:histone demethylase jarid1a;
PDBTitle: nmr structure of the arid domain from the histone h3k42 demethylase rbp2
|
|
|