| 1 |
|
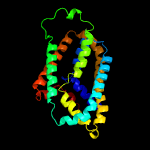
PDB 5vkv chain A
Region: 15 - 259
Aligned: 220
Modelled: 227
Confidence: 100.0%
Identity: 34%
PDB header:oxidoreductase
Chain: A: PDB Molecule:cytochrome c-type biogenesis protein ccda;
PDBTitle: solution nmr structure of the membrane electron transporter ccda
Phyre2
| 2 |
|
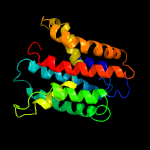
PDB 2n4x chain A
Region: 19 - 240
Aligned: 186
Modelled: 194
Confidence: 100.0%
Identity: 18%
PDB header:membrane protein
Chain: A: PDB Molecule:cytochrome c-type biogenesis protein (ccda);
PDBTitle: structure of the transmembrane electron transporter ccda
Phyre2
| 3 |
|
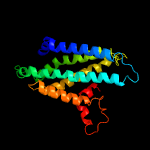
PDB 5njg chain B
Region: 60 - 258
Aligned: 195
Modelled: 199
Confidence: 20.3%
Identity: 10%
PDB header:transport protein
Chain: B: PDB Molecule:atp-binding cassette sub-family g member 2;
PDBTitle: structure of an abc transporter: part of the structure that could be2 built de novo
Phyre2
| 4 |
|
PDB 4zjv chain D
Region: 30 - 37
Aligned: 8
Modelled: 8
Confidence: 15.0%
Identity: 50%
PDB header:transferase/inhibitor
Chain: D: PDB Molecule:erbb receptor feedback inhibitor 1;
PDBTitle: crystal structure of egfr kinase domain in complex with mitogen-2 inducible gene 6 protein
Phyre2
| 5 |
|
PDB 1v54 chain L
Region: 182 - 199
Aligned: 18
Modelled: 18
Confidence: 13.6%
Identity: 17%
Fold: Single transmembrane helix
Superfamily: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)
Family: Mitochondrial cytochrome c oxidase subunit VIIc (aka VIIIa)
Phyre2
| 6 |
|
PDB 2y69 chain Y
Region: 182 - 199
Aligned: 18
Modelled: 18
Confidence: 10.8%
Identity: 17%
PDB header:electron transport
Chain: Y: PDB Molecule:cytochrome c oxidase subunit 7c;
PDBTitle: bovine heart cytochrome c oxidase re-refined with molecular oxygen
Phyre2
| 7 |
|
PDB 5do7 chain B
Region: 47 - 258
Aligned: 208
Modelled: 212
Confidence: 9.5%
Identity: 10%
PDB header:transport protein
Chain: B: PDB Molecule:atp-binding cassette sub-family g member 8;
PDBTitle: crystal structure of the human sterol transporter abcg5/abcg8
Phyre2
| 8 |
|
PDB 1xyr chain 6
Region: 30 - 35
Aligned: 6
Modelled: 6
Confidence: 9.2%
Identity: 67%
PDB header:virus
Chain: 6: PDB Molecule:genome polyprotein, coat protein vp3;
PDB Fragment:residues 620-630
PDBTitle: poliovirus 135s cell entry intermediate
Phyre2
| 9 |
|
PDB 6mjp chain F
Region: 25 - 138
Aligned: 94
Modelled: 94
Confidence: 7.5%
Identity: 10%
PDB header:lipid transport
Chain: F: PDB Molecule:fig000988: predicted permease;
PDBTitle: lptb(e163q)fgc from vibrio cholerae
Phyre2
| 10 |
|
PDB 4p79 chain A
Region: 177 - 251
Aligned: 75
Modelled: 75
Confidence: 6.4%
Identity: 17%
PDB header:cell adhesion
Chain: A: PDB Molecule:claudin-15;
PDBTitle: crystal structure of mouse claudin-15
Phyre2
| 11 |
|
PDB 2hg5 chain D
Region: 222 - 249
Aligned: 28
Modelled: 28
Confidence: 6.2%
Identity: 11%
PDB header:membrane protein
Chain: D: PDB Molecule:kcsa channel;
PDBTitle: cs+ complex of a k channel with an amide to ester substitution in the2 selectivity filter
Phyre2
| 12 |
|
PDB 1eak chain A domain 5
Region: 30 - 35
Aligned: 6
Modelled: 6
Confidence: 5.1%
Identity: 67%
Fold: Kringle-like
Superfamily: Kringle-like
Family: Fibronectin type II module
Phyre2