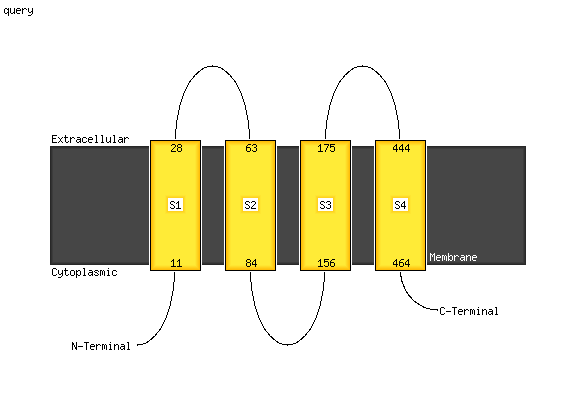
1 d1y5ic1
68.9
25
Fold: Heme-binding four-helical bundleSuperfamily: Respiratory nitrate reductase 1 gamma chainFamily: Respiratory nitrate reductase 1 gamma chain
2 c5lm4A_
63.7
19
PDB header: transport proteinChain: A: PDB Molecule: excitatory amino acid transporter 1,neutral amino acidPDBTitle: structure of the thermostalilized eaat1 cryst-ii mutant in complex2 with l-asp and the allosteric inhibitor ucph101
3 d1wjwa_
42.0
19
Fold: TBP-likeSuperfamily: Phosphoglucomutase, C-terminal domainFamily: Phosphoglucomutase, C-terminal domain
4 c6oceB_
28.2
15
PDB header: transport proteinChain: B: PDB Molecule: stress-gated cation channel 1.2;PDBTitle: structure of the rice hyperosmolality-gated ion channel osca1.2
5 c2k21A_
18.0
25
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily ePDBTitle: nmr structure of human kcne1 in lmpg micelles at ph 6.0 and2 40 degree c
6 c2ndjA_
15.8
17
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 3;PDBTitle: structural basis for kcne3 and estrogen modulation of the kcnq12 channel
7 c2k9yA_
15.6
27
PDB header: transferaseChain: A: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
8 c2k9yB_
15.6
27
PDB header: transferaseChain: B: PDB Molecule: ephrin type-a receptor 2;PDBTitle: epha2 dimeric structure in the lipidic bicelle at ph 5.0
9 c1f20A_
15.3
12
PDB header: oxidoreductaseChain: A: PDB Molecule: nitric-oxide synthase;PDBTitle: crystal structure of rat neuronal nitric-oxide synthase fad/nadp+2 domain at 1.9a resolution.
10 c1bhbA_
12.4
26
PDB header: photoreceptorChain: A: PDB Molecule: bacteriorhodopsin;PDBTitle: three-dimensional structure of (1-71) bacterioopsin2 solubilized in methanol-chloroform and sds micelles3 determined by 15n-1h heteronuclear nmr spectroscopy
11 c5ochF_
11.8
22
PDB header: hydrolaseChain: F: PDB Molecule: atp-binding cassette sub-family b member 8, mitochondrial;PDBTitle: the crystal structure of human abcb8 in an outward-facing state
12 c2m0qA_
11.5
21
PDB header: membrane proteinChain: A: PDB Molecule: potassium voltage-gated channel subfamily e member 2;PDBTitle: solution nmr analysis of intact kcne2 in detergent micelles2 demonstrate a straight transmembrane helix
13 c6nd1E_
11.3
13
PDB header: protein transportChain: E: PDB Molecule: translocation protein sec66;PDBTitle: cryoem structure of the sec complex from yeast
14 c5ws4A_
11.0
15
PDB header: membrane proteinChain: A: PDB Molecule: macrolide export atp-binding/permease protein macb;PDBTitle: crystal structure of tripartite-type abc transporter macb from2 acinetobacter baumannii
15 c2kluA_
10.4
32
PDB header: immune system, membrane proteinChain: A: PDB Molecule: t-cell surface glycoprotein cd4;PDBTitle: nmr structure of the transmembrane and cytoplasmic domains2 of human cd4
16 c2ks1B_
9.9
28
PDB header: transferaseChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: heterodimeric association of transmembrane domains of erbb1 and erbb22 receptors enabling kinase activation
17 c5a43B_
9.5
33
PDB header: transport proteinChain: B: PDB Molecule: putative fluoride ion transporter crcb;PDBTitle: crystal structure of a dual topology fluoride ion channel.
18 c2m1jA_
9.4
23
PDB header: unknown functionChain: A: PDB Molecule: prion-like protein doppel;PDBTitle: ovine doppel signal peptide (1-30)
19 c5gasN_
8.8
14
PDB header: hydrolaseChain: N: PDB Molecule: archaeal/vacuolar-type h+-atpase subunit i;PDBTitle: thermus thermophilus v/a-atpase, conformation 2
20 c6adqP_
8.3
30
PDB header: electron transportChain: P: PDB Molecule: prokaryotic respiratory supercomplex associate factor 1PDBTitle: respiratory complex ciii2civ2sod2 from mycobacterium smegmatis
21 c6gctA_
not modelled
8.1
21
PDB header: membrane proteinChain: A: PDB Molecule: neutral amino acid transporter b(0);PDBTitle: cryo-em structure of the human neutral amino acid transporter asct2
22 c2m20B_
not modelled
8.0
28
PDB header: signaling proteinChain: B: PDB Molecule: epidermal growth factor receptor;PDBTitle: egfr transmembrane - juxtamembrane (tm-jm) segment in bicelles: md2 guided nmr refined structure.
23 c3j1rP_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: P: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
24 c3j1rE_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: E: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
25 c3j1rF_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: F: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
26 c3j1rB_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: B: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
27 c3j1rS_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: S: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
28 c3j1rO_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: O: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
29 c3j1rM_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: M: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
30 c3j1rR_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: R: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
31 c3j1rH_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: H: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
32 c3j1rI_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: I: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
33 c3j1rG_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: G: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
34 c3j1rL_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: L: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
35 c3j1rN_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: N: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
36 c3j1rC_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: C: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
37 c3j1rQ_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: Q: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
38 c3j1rU_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: U: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
39 c3j1rA_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: A: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
40 c3j1rD_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: D: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
41 c3j1rJ_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: J: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
42 c3j1rK_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: K: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
43 c3j1rT_
not modelled
7.9
44
PDB header: cell adhesion, structural proteinChain: T: PDB Molecule: archaeal adhesion filament core;PDBTitle: filaments from ignicoccus hospitalis show diversity of packing in2 proteins containing n-terminal type iv pilin helices
44 d1xrda1
not modelled
7.7
27
Fold: Light-harvesting complex subunitsSuperfamily: Light-harvesting complex subunitsFamily: Light-harvesting complex subunits
45 c1xrdA_
not modelled
7.7
27
PDB header: membrane proteinChain: A: PDB Molecule: light-harvesting protein b-880, alpha chain;PDBTitle: light-harvesting complex 1 alfa subunit from wild-type2 rhodospirillum rubrum
46 d1qhda2
not modelled
7.5
39
Fold: Viral protein domainSuperfamily: Viral protein domainFamily: Top domain of virus capsid protein
47 c1afoB_
not modelled
7.4
22
PDB header: integral membrane proteinChain: B: PDB Molecule: glycophorin a;PDBTitle: dimeric transmembrane domain of human glycophorin a, nmr,2 20 structures
48 c2mpnB_
not modelled
7.3
15
PDB header: membrane proteinChain: B: PDB Molecule: inner membrane protein ygap;PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
49 c2mpnA_
not modelled
7.3
15
PDB header: membrane proteinChain: A: PDB Molecule: inner membrane protein ygap;PDBTitle: 3d nmr structure of the transmembrane domain of the full-length inner2 membrane protein ygap from escherichia coli
50 c6et5b_
not modelled
7.0
23
PDB header: photosynthesisChain: B: PDB Molecule: PDBTitle: reaction centre light harvesting complex 1 from blc. virids
51 c3wmmY_
not modelled
6.9
32
PDB header: photosynthesisChain: Y: PDB Molecule: lh1 alpha polypeptide;PDBTitle: crystal structure of the lh1-rc complex from thermochromatium tepidum2 in c2 form
52 c5mkkB_
not modelled
5.9
16
PDB header: transport proteinChain: B: PDB Molecule: multidrug resistance abc transporter atp-binding andPDBTitle: crystal structure of the heterodimeric abc transporter tmrab, a2 homolog of the antigen translocation complex tap
53 c5eh4C_
not modelled
5.9
24
PDB header: membrane proteinChain: C: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane dimer in lipidic2 cubic phase
54 c5eh4A_
not modelled
5.9
24
PDB header: membrane proteinChain: A: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane dimer in lipidic2 cubic phase
55 c5eh4B_
not modelled
5.9
24
PDB header: membrane proteinChain: B: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane dimer in lipidic2 cubic phase
56 c5eh4D_
not modelled
5.9
24
PDB header: membrane proteinChain: D: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane dimer in lipidic2 cubic phase
57 c5eh6A_
not modelled
5.9
24
PDB header: membrane proteinChain: A: PDB Molecule: glycophorin-a;PDBTitle: crystal structure of the glycophorin a transmembrane monomer in2 lipidic cubic phase
58 c2mc0A_
not modelled
5.7
27
PDB header: transcription activator/antibioticChain: A: PDB Molecule: hth-type transcriptional activator tipa;PDBTitle: structural basis of a thiopeptide antibiotic multidrug resistance2 system from streptomyces lividans:nosiheptide in complex with tipas
59 c2na9A_
not modelled
5.7
18
PDB header: signaling proteinChain: A: PDB Molecule: cytokine receptor common subunit beta;PDBTitle: transmembrane structure of the p441a mutant of the cytokine receptor2 common subunit beta
60 c2na8A_
not modelled
5.7
18
PDB header: membrane proteinChain: A: PDB Molecule: cytokine receptor common subunit beta;PDBTitle: transmembrane structure of the cytokine receptor common subunit beta
61 c3wvfA_
not modelled
5.6
15
PDB header: chaperoneChain: A: PDB Molecule: membrane protein insertase yidc;PDBTitle: crystal structure of yidc from escherichia coli
62 c5sv1Y_
not modelled
5.5
30
PDB header: transport proteinChain: Y: PDB Molecule: biopolymer transport protein exbd;PDBTitle: structure of the exbb/exbd complex from e. coli at ph 4.5
63 c3iykA_
not modelled
5.5
43
PDB header: virusChain: A: PDB Molecule: vp5;PDBTitle: bluetongue virus structure reveals a sialic acid binding domain,2 amphipathic helices and a central coiled coil in the outer capsid3 proteins
64 c2n2aA_
not modelled
5.4
21
PDB header: membrane proteinChain: A: PDB Molecule: receptor tyrosine-protein kinase erbb-2;PDBTitle: spatial structure of her2/erbb2 dimeric transmembrane domain in the2 presence of cytoplasmic juxtamembrane domains
65 c5sv1Z_
not modelled
5.4
30
PDB header: transport proteinChain: Z: PDB Molecule: biopolymer transport protein exbd;PDBTitle: structure of the exbb/exbd complex from e. coli at ph 4.5
66 c5u1dA_
not modelled
5.3
15
PDB header: transport proteinChain: A: PDB Molecule: antigen peptide transporter 1;PDBTitle: cryo-em structure of the human tap atp-binding cassette transporter
67 c6f2wA_
not modelled
5.3
4
PDB header: transport proteinChain: A: PDB Molecule: putative amino acid/polyamine transport protein;PDBTitle: bacterial asc transporter crystal structure in open to in conformation
68 c4pe5A_
not modelled
5.2
20
PDB header: transport proteinChain: A: PDB Molecule: glutamate receptor ionotropic, nmda 1;PDBTitle: crystal structure of glun1a/glun2b nmda receptor ion channel