| 1 |
|
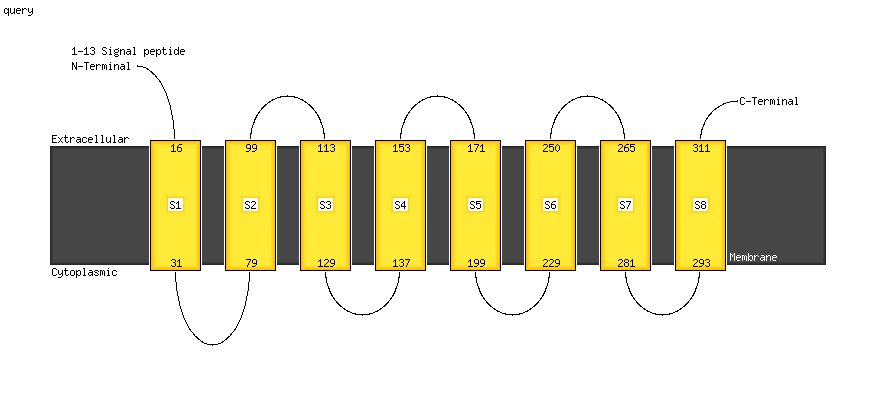
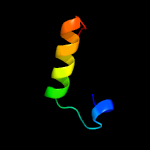
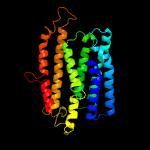
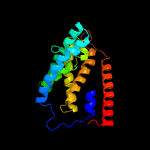
PDB 5ir6 chain B
Region: 107 - 308
Aligned: 186
Modelled: 186
Confidence: 46.0%
Identity: 9%
PDB header:oxidoreductase
Chain: B: PDB Molecule:bd-type quinol oxidase subunit ii;
PDBTitle: the structure of bd oxidase from geobacillus thermodenitrificans
Phyre2
| 2 |
|
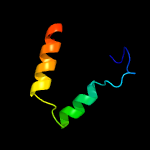
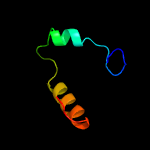
PDB 3ayg chain A
Region: 157 - 322
Aligned: 166
Modelled: 166
Confidence: 31.9%
Identity: 11%
PDB header:oxidoreductase
Chain: A: PDB Molecule:nitric oxide reductase;
PDBTitle: crystal structure of nitric oxide reductase complex with hqno
Phyre2
| 3 |
|
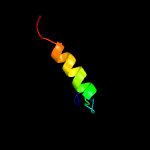
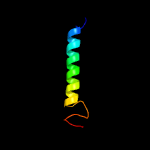
PDB 5azc chain A
Region: 238 - 283
Aligned: 46
Modelled: 46
Confidence: 29.5%
Identity: 22%
PDB header:transferase
Chain: A: PDB Molecule:prolipoprotein diacylglyceryl transferase;
PDBTitle: crystal structure of escherichia coli lgt in complex with2 phosphatidylglycerol
Phyre2
| 4 |
|
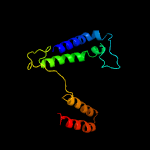
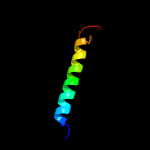
PDB 6o7u chain A
Region: 147 - 309
Aligned: 151
Modelled: 163
Confidence: 28.9%
Identity: 13%
PDB header:membrane protein
Chain: A: PDB Molecule:
PDBTitle: saccharomyces cerevisiae v-atpase stv1-vo
Phyre2
| 5 |
|
PDB 3mk7 chain K
Region: 68 - 308
Aligned: 213
Modelled: 239
Confidence: 25.4%
Identity: 15%
PDB header:oxidoreductase
Chain: K: PDB Molecule:cytochrome c oxidase, cbb3-type, subunit n;
PDBTitle: the structure of cbb3 cytochrome oxidase
Phyre2
| 6 |
|
PDB 6gcs chain X
Region: 241 - 266
Aligned: 26
Modelled: 26
Confidence: 19.8%
Identity: 23%
PDB header:oxidoreductase
Chain: X: PDB Molecule:nuxm subunit;
PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
Phyre2
| 7 |
|
PDB 3ol4 chain B
Region: 212 - 239
Aligned: 28
Modelled: 28
Confidence: 19.6%
Identity: 18%
PDB header:unknown function
Chain: B: PDB Molecule:putative uncharacterized protein;
PDBTitle: crystal structure of a putative uncharacterized protein from2 mycobacterium smegmatis, an ortholog of rv0543c
Phyre2
| 8 |
|
PDB 2lky chain A
Region: 197 - 239
Aligned: 43
Modelled: 43
Confidence: 19.3%
Identity: 9%
PDB header:structural genomics, unknown function
Chain: A: PDB Molecule:uncharacterized protein;
PDBTitle: solution structure of msmeg_1053, the second duf3349 annotated protein2 in the genome of mycobacterium smegmatis, seattle structural genomics3 center for infectious disease target mysma.17112.b
Phyre2
| 9 |
|
PDB 2m0n chain A
Region: 212 - 239
Aligned: 28
Modelled: 28
Confidence: 16.7%
Identity: 25%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution structure of a duf3349 annotated protein from mycobacterium2 abscessus, mab_3403c. seattle structural genomics center for3 infectious disease target myaba.17112.a.a2
Phyre2
| 10 |
|
PDB 6o7x chain A
Region: 181 - 309
Aligned: 117
Modelled: 129
Confidence: 14.9%
Identity: 14%
PDB header:membrane protein
Chain: A: PDB Molecule:vacuolar atp synthase catalytic subunit a;
PDBTitle: saccharomyces cerevisiae v-atpase stv1-v1vo state 3
Phyre2
| 11 |
|
PDB 3o0r chain B
Region: 13 - 307
Aligned: 243
Modelled: 264
Confidence: 14.0%
Identity: 12%
PDB header:immune system/oxidoreductase
Chain: B: PDB Molecule:nitric oxide reductase subunit b;
PDBTitle: crystal structure of nitric oxide reductase from pseudomonas2 aeruginosa in complex with antibody fragment
Phyre2
| 12 |
|
PDB 2kvc chain A
Region: 197 - 239
Aligned: 43
Modelled: 43
Confidence: 12.8%
Identity: 14%
PDB header:unknown function
Chain: A: PDB Molecule:putative uncharacterized protein;
PDBTitle: solution structure of the mycobacterium tuberculosis protein rv0543c,2 a member of the duf3349 superfamily. seattle structural genomics3 center for infectious disease target mytud.17112.a
Phyre2
| 13 |
|
PDB 5xnm chain J
Region: 289 - 324
Aligned: 32
Modelled: 36
Confidence: 9.5%
Identity: 28%
PDB header:membrane protein
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: structure of unstacked c2s2m2-type psii-lhcii supercomplex from pisum2 sativum
Phyre2
| 14 |
|
PDB 3jcu chain J
Region: 289 - 324
Aligned: 32
Modelled: 36
Confidence: 9.3%
Identity: 28%
PDB header:membrane protein
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: cryo-em structure of spinach psii-lhcii supercomplex at 3.2 angstrom2 resolution
Phyre2
| 15 |
|
PDB 1u7g chain A
Region: 28 - 291
Aligned: 202
Modelled: 208
Confidence: 7.1%
Identity: 18%
Fold: Ammonium transporter
Superfamily: Ammonium transporter
Family: Ammonium transporter
Phyre2
| 16 |
|
PDB 2axt chain J domain 1
Region: 319 - 324
Aligned: 6
Modelled: 6
Confidence: 6.6%
Identity: 67%
Fold: Single transmembrane helix
Superfamily: Photosystem II reaction center protein J, PsbJ
Family: PsbJ-like
Phyre2
| 17 |
|
PDB 3a0h chain J
Region: 319 - 324
Aligned: 6
Modelled: 6
Confidence: 6.6%
Identity: 67%
PDB header:electron transport
Chain: J: PDB Molecule:photosystem ii reaction center protein j;
PDBTitle: crystal structure of i-substituted photosystem ii complex
Phyre2