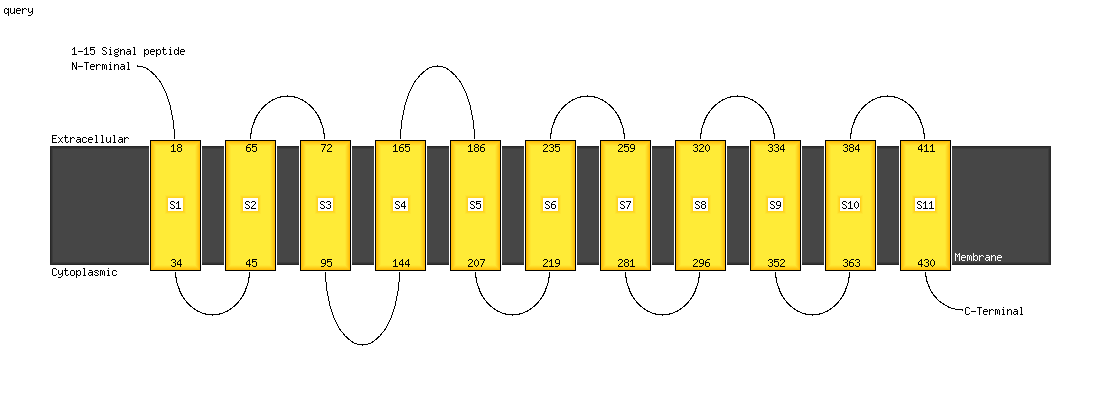
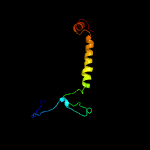
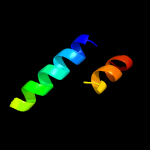
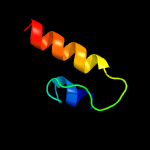
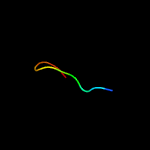
1 c1yewI_
83.9
19
PDB header: oxidoreductase, membrane proteinChain: I: PDB Molecule: particulate methane monooxygenase, b subunit;PDBTitle: crystal structure of particulate methane monooxygenase
2 c3rfrI_
83.0
19
PDB header: oxidoreductaseChain: I: PDB Molecule: pmob;PDBTitle: crystal structure of particulate methane monooxygenase (pmmo) from2 methylocystis sp. strain m
3 c3rgbA_
80.9
19
PDB header: oxidoreductaseChain: A: PDB Molecule: methane monooxygenase subunit b2;PDBTitle: crystal structure of particulate methane monooxygenase from2 methylococcus capsulatus (bath)
4 c4uisB_
51.5
15
PDB header: hydrolaseChain: B: PDB Molecule: gamma-secretase;PDBTitle: the cryoem structure of human gamma-secretase complex
5 c5a63D_
33.1
21
PDB header: hydrolaseChain: D: PDB Molecule: gamma-secretase subunit pen-2;PDBTitle: cryo-em structure of the human gamma-secretase complex at 3.4 angstrom2 resolution.
6 c3w3fB_
26.5
29
PDB header: biosynthetic proteinChain: B: PDB Molecule: uncharacterized protein blr2150;PDBTitle: crystal structure of ent-kaurene synthase bjks from bradyrhizobium2 japonicum
7 d2p7tc1
25.1
47
Fold: Voltage-gated potassium channelsSuperfamily: Voltage-gated potassium channelsFamily: Voltage-gated potassium channels
8 c4pw1B_
20.0
67
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: uncharacterized protein;PDBTitle: crystal structure of a protein with unknown function (clolep_02462)2 from clostridium leptum dsm 753 at 2.10 a resolution
9 c6h9iB_
18.9
55
PDB header: rna binding proteinChain: B: PDB Molecule: csf5;PDBTitle: csf5, crispr-cas type iv cas6 crrna endonuclease
10 c3chxE_
18.7
23
PDB header: membrane proteinChain: E: PDB Molecule: pmob;PDBTitle: crystal structure of methylosinus trichosporium ob3b particulate2 methane monooxygenase (pmmo)
11 c3nglA_
15.8
100
PDB header: oxidoreductase, hydrolaseChain: A: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of bifunctional 5,10-methylenetetrahydrofolate2 dehydrogenase / cyclohydrolase from thermoplasma acidophilum
12 c5nhsB_
15.5
100
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional protein fold;PDBTitle: the crystal structure of xanthomonas albilineans n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
13 c4o65A_
15.4
38
PDB header: oxidoreductaseChain: A: PDB Molecule: putative archaeal ammonia monooxygenase subunit b;PDBTitle: crystal structure of the cupredoxin domain of amob from nitrosocaldus2 yellowstonii
14 c1a4iB_
14.5
100
PDB header: oxidoreductaseChain: B: PDB Molecule: methylenetetrahydrofolate dehydrogenase /PDBTitle: human tetrahydrofolate dehydrogenase / cyclohydrolase
15 d1kjna_
14.4
36
Fold: Hypothetical protein MTH777 (MT0777)Superfamily: Hypothetical protein MTH777 (MT0777)Family: Hypothetical protein MTH777 (MT0777)
16 c2c2xB_
14.4
86
PDB header: oxidoreductaseChain: B: PDB Molecule: methylenetetrahydrofolate dehydrogenase-PDBTitle: three dimensional structure of bifunctional methylenetetrahydrofolate2 dehydrogenase-cyclohydrolase from mycobacterium tuberculosis
17 c5zf1A_
14.2
100
PDB header: oxidoreductaseChain: A: PDB Molecule: 5,10-methylenetetrahydrofolate dehydrogenase;PDBTitle: molecular structure of a novel 5,10-methylenetetrahydrofolate2 dehydrogenase from the silkworm, bombyx mori
18 c1b0aA_
14.0
100
PDB header: oxidoreductase,hydrolaseChain: A: PDB Molecule: protein (fold bifunctional protein);PDBTitle: 5,10, methylene-tetrahydropholate2 dehydrogenase/cyclohydrolase from e coli.
19 c5z70B_
13.5
20
PDB header: hydrolaseChain: B: PDB Molecule: oleate hydratase;PDBTitle: crystal strucure of oleate hydratase from stenotrophomonas sp. kctc2 12332
20 c4cjxA_
13.0
100
PDB header: oxidoreductaseChain: A: PDB Molecule: c-1-tetrahydrofolate synthase, cytoplasmic, putative;PDBTitle: the crystal structure of trypanosoma brucei n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)3 complexed with nadp cofactor and inhibitor
21 c4b4uB_
not modelled
12.8
100
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of acinetobacter baumannii n5,2 n10-methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)3 complexed with nadp cofactor
22 c3p2oB_
not modelled
12.6
100
PDB header: oxidoreductase, hydrolaseChain: B: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
23 c4a5oB_
not modelled
12.5
100
PDB header: oxidoreductaseChain: B: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of pseudomonas aeruginosa n5, n10-2 methylenetetrahydrofolate dehydrogenase-cyclohydrolase (fold)
24 c3l07B_
not modelled
12.5
100
PDB header: oxidoreductase,hydrolaseChain: B: PDB Molecule: bifunctional protein fold;PDBTitle: methylenetetrahydrofolate dehydrogenase/methenyltetrahydrofolate2 cyclohydrolase, putative bifunctional protein fold from francisella3 tularensis.
25 c3p2oA_
not modelled
12.3
100
PDB header: oxidoreductase, hydrolaseChain: A: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of fold bifunctional protein from campylobacter2 jejuni
26 c5tc4A_
not modelled
12.3
100
PDB header: oxidoreductaseChain: A: PDB Molecule: bifunctional methylenetetrahydrofolatePDBTitle: crystal structure of human mitochondrial methylenetetrahydrofolate2 dehydrogenase-cyclohydrolase (mthfd2) in complex with ly345899 and3 cofactors
27 d1kshb_
not modelled
12.3
53
Fold: Immunoglobulin-like beta-sandwichSuperfamily: E set domainsFamily: RhoGDI-like
28 d1b0aa1
not modelled
11.7
100
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
29 d1xtda2
not modelled
11.7
46
Fold: OB-foldSuperfamily: Nucleic acid-binding proteinsFamily: Cold shock DNA-binding domain-like
30 c6apeA_
not modelled
11.7
86
PDB header: oxidoreductase, hydrolaseChain: A: PDB Molecule: bifunctional protein fold;PDBTitle: crystal structure of bifunctional protein fold from helicobacter2 pylori
31 c4a26B_
not modelled
11.5
100
PDB header: oxidoreductaseChain: B: PDB Molecule: putative c-1-tetrahydrofolate synthase, cytoplasmic;PDBTitle: the crystal structure of leishmania major n5,n10-2 methylenetetrahydrofolate dehydrogenase/cyclohydrolase
32 c4ak2A_
not modelled
11.0
29
PDB header: heparin-binding proteinChain: A: PDB Molecule: bt_4661;PDBTitle: structure of bt4661, a suse-like surface located polysaccharide2 binding protein from the bacteroides thetaiotaomicron heparin3 utilisation locus
33 d1a34a_
not modelled
10.6
43
Fold: Nucleoplasmin-like/VP (viral coat and capsid proteins)Superfamily: Satellite virusesFamily: Satellite viruses
34 c5odoA_
not modelled
10.0
20
PDB header: lyaseChain: A: PDB Molecule: isomerase;PDBTitle: crystal structure of the oleate hydratase of rhodococcus erythropolis
35 c4uirB_
not modelled
9.9
25
PDB header: lyaseChain: B: PDB Molecule: oleate hydratase;PDBTitle: structure of oleate hydratase from elizabethkingia2 meningoseptica
36 c6gcs3_
not modelled
9.8
20
PDB header: oxidoreductaseChain: 3: PDB Molecule: nd3 subunit (nu3m);PDBTitle: cryo-em structure of respiratory complex i from yarrowia lipolytica
37 d1a4ia1
not modelled
9.6
100
Fold: NAD(P)-binding Rossmann-fold domainsSuperfamily: NAD(P)-binding Rossmann-fold domainsFamily: Aminoacid dehydrogenase-like, C-terminal domain
38 c2mi2A_
not modelled
9.4
30
PDB header: transport proteinChain: A: PDB Molecule: sec-independent protein translocase protein tatb;PDBTitle: solution structure of the e. coli tatb protein in dpc micelles
39 c4gokG_
not modelled
9.2
47
PDB header: signaling proteinChain: G: PDB Molecule: protein unc-119 homolog a;PDBTitle: the crystal structure of arl2gppnhp in complex with unc119a
40 c2kr6A_
not modelled
9.2
19
PDB header: hydrolaseChain: A: PDB Molecule: presenilin-1;PDBTitle: solution structure of presenilin-1 ctf subunit
41 c2mckA_
not modelled
8.9
43
PDB header: hydrolaseChain: A: PDB Molecule: polyprotein;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for murine2 norovirus cr6 ns1/2 protein
42 c3rkoE_
not modelled
8.1
28
PDB header: oxidoreductaseChain: E: PDB Molecule: nadh-quinone oxidoreductase subunit a;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
43 c3rkoA_
not modelled
8.1
28
PDB header: oxidoreductaseChain: A: PDB Molecule: nadh-quinone oxidoreductase subunit a;PDBTitle: crystal structure of the membrane domain of respiratory complex i from2 e. coli at 3.0 angstrom resolution
44 c2mcdA_
not modelled
7.4
43
PDB header: hydrolaseChain: A: PDB Molecule: murine norovirus 1;PDBTitle: backbone 1h, 13c, and 15n chemical shift assignments for murine2 norovirus ns1/2 d94e mutant
45 c2k5cA_
not modelled
7.3
17
PDB header: metal binding proteinChain: A: PDB Molecule: uncharacterized protein pf0385;PDBTitle: nmr structure for pf0385
46 c4m5bA_
not modelled
7.2
20
PDB header: membrane proteinChain: A: PDB Molecule: cobalamin biosynthesis protein cbim;PDBTitle: crystal structure of an truncated transition metal transporter
47 c4m5cA_
not modelled
7.2
20
PDB header: membrane proteinChain: A: PDB Molecule: cobalamin biosynthesis protein cbim;PDBTitle: crystal structure of an truncated transition metal transporter
48 c6mwqB_
not modelled
7.2
36
PDB header: lyase/fluorescent proteinChain: B: PDB Molecule: darpin, muscle-type aldolase chimeric fusion;PDBTitle: single particle cryoem structure of a darpin-aldolase platform in2 complex with gfp
49 c1n93X_
not modelled
7.1
30
PDB header: viral proteinChain: X: PDB Molecule: p40 nucleoprotein;PDBTitle: crystal structure of the borna disease virus nucleoprotein
50 d1n93x_
not modelled
7.1
30
Fold: P40 nucleoproteinSuperfamily: P40 nucleoproteinFamily: P40 nucleoprotein
51 c3kihC_
not modelled
6.9
40
PDB header: sugar binding proteinChain: C: PDB Molecule: 5-bladed beta-propeller lectin;PDBTitle: the crystal structures of two fragments truncated from 5-bladed beta-2 propeller lectin, tachylectin-2 (lib2-d2-15)
52 c5d6hB_
not modelled
6.8
50
PDB header: chaperone/protein transportChain: B: PDB Molecule: csua/b;PDBTitle: crystal structure of csuc-csua/b chaperone-major subunit pre-assembly2 complex from csu biofilm-mediating pili of acinetobacter baumannii
53 d1ro7a_
not modelled
6.7
37
Fold: Alpha-2,3/8-sialyltransferase CstIISuperfamily: Alpha-2,3/8-sialyltransferase CstIIFamily: Alpha-2,3/8-sialyltransferase CstII
54 c3qheB_
not modelled
6.6
100
PDB header: signaling protein/splicingChain: B: PDB Molecule: kh domain-containing, rna-binding, signal transduction-PDBTitle: crystal structure of the complex between the armadillo repeat domain2 of adenomatous polyposis coli and the tyrosine-rich domain of sam68
55 d1ztda1
not modelled
6.5
25
Fold: RNase III domain-likeSuperfamily: RNase III domain-likeFamily: PF0609-like
56 c2ru8A_
not modelled
6.5
25
PDB header: replicationChain: A: PDB Molecule: primosomal protein 1;PDBTitle: dnat c-terminal domain
57 d1rzhh1
not modelled
6.5
71
Fold: PRC-barrel domainSuperfamily: PRC-barrel domainFamily: Photosynthetic reaction centre, H-chain, cytoplasmic domain
58 d1droa_
not modelled
6.4
50
Fold: PH domain-like barrelSuperfamily: PH domain-likeFamily: Pleckstrin-homology domain (PH domain)
59 c3wvbA_
not modelled
6.4
67
PDB header: hydrolaseChain: A: PDB Molecule: upf0254 protein mj1251;PDBTitle: hcgf from methanocaldococcus jannaschii
60 c5djbD_
not modelled
6.4
21
PDB header: structural proteinChain: D: PDB Molecule: microcompartments protein;PDBTitle: structure of the haliangium ochraceum bmc-h shell protein
61 c2do6A_
not modelled
6.4
23
PDB header: ligaseChain: A: PDB Molecule: e3 ubiquitin-protein ligase cbl-b;PDBTitle: solution structure of rsgi ruh-065, a uba domain from human2 cdna
62 d1jmxg_
not modelled
6.3
55
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Quinohemoprotein amine dehydrogenase C chainFamily: Quinohemoprotein amine dehydrogenase C chain
63 d2c42a2
not modelled
6.3
33
Fold: Thiamin diphosphate-binding fold (THDP-binding)Superfamily: Thiamin diphosphate-binding fold (THDP-binding)Family: PFOR PP module
64 c5wsnC_
not modelled
6.2
16
PDB header: virusChain: C: PDB Molecule: e protein;PDBTitle: structure of japanese encephalitis virus
65 c6r7tB_
not modelled
6.2
32
PDB header: immune systemChain: B: PDB Molecule: melanoma-associated antigen b1;PDBTitle: crystal structure of human melanoma-associated antigen b1 (mageb1) in2 complex with nanobody
66 c3gczA_
not modelled
6.1
47
PDB header: transferaseChain: A: PDB Molecule: polyprotein;PDBTitle: yokose virus methyltransferase in complex with adomet
67 d1pbyc_
not modelled
5.9
45
Fold: Non-globular all-alpha subunits of globular proteinsSuperfamily: Quinohemoprotein amine dehydrogenase C chainFamily: Quinohemoprotein amine dehydrogenase C chain
68 c1lt1G_
not modelled
5.9
37
PDB header: de novo proteinChain: G: PDB Molecule: l13g-df1;PDBTitle: sliding helix induced change of coordination geometry in a2 model di-mn(ii) protein
69 c6e3yP_
not modelled
5.8
62
PDB header: signaling proteinChain: P: PDB Molecule: calcitonin gene-related peptide 1;PDBTitle: cryo-em structure of the active, gs-protein complexed, human cgrp2 receptor
70 c2ekkA_
not modelled
5.7
18
PDB header: protein bindingChain: A: PDB Molecule: uba domain from e3 ubiquitin-protein ligasePDBTitle: solution structure of ruh-074, a human uba domain
71 c4ou6A_
not modelled
5.7
25
PDB header: replication/dnaChain: A: PDB Molecule: primosomal protein 1;PDBTitle: crystal structure of dnat84-153-dt10 ssdna complex form 1
72 c2kglA_
not modelled
5.4
14
PDB header: chaperoneChain: A: PDB Molecule: mesoderm development candidate 2;PDBTitle: nmr solution structure of mesd
73 c3eeqB_
not modelled
5.4
16
PDB header: structural genomics, unknown functionChain: B: PDB Molecule: putative cobalamin biosynthesis protein g homolog;PDBTitle: crystal structure of a putative cobalamin biosynthesis protein g2 homolog from sulfolobus solfataricus
74 d1igna2
not modelled
5.4
43
Fold: DNA/RNA-binding 3-helical bundleSuperfamily: Homeodomain-likeFamily: DNA-binding domain of rap1
75 d1muka_
not modelled
5.4
39
Fold: DNA/RNA polymerasesSuperfamily: DNA/RNA polymerasesFamily: RNA-dependent RNA-polymerase
76 c2wa0A_
not modelled
5.4
32
PDB header: immune systemChain: A: PDB Molecule: melanoma-associated antigen 4;PDBTitle: crystal structure of the human magea4
77 d2axtk1
not modelled
5.4
47
Fold: Single transmembrane helixSuperfamily: Photosystem II reaction center protein K, PsbKFamily: PsbK-like
78 d1ciya3
not modelled
5.3
20
Fold: Toxins' membrane translocation domainsSuperfamily: delta-Endotoxin (insectocide), N-terminal domainFamily: delta-Endotoxin (insectocide), N-terminal domain
79 c4djhA_
not modelled
5.3
11
PDB header: hormone receptor/antagonistChain: A: PDB Molecule: kappa-type opioid receptor, lysozyme;PDBTitle: structure of the human kappa opioid receptor in complex with jdtic
80 c1w99A_
not modelled
5.2
20
PDB header: toxinChain: A: PDB Molecule: pesticidial crystal protein cry4ba;PDBTitle: mosquito-larvicidal toxin cry4ba from bacillus thuringiensis ssp.2 israelensis
81 c1kzvA_
not modelled
5.2
29
PDB header: viral proteinChain: A: PDB Molecule: vpr protein;PDBTitle: structure of human immunodeficiency virus type 1 vpr(34-51)2 peptide in chloroform methanol
82 c1kztA_
not modelled
5.2
29
PDB header: viral proteinChain: A: PDB Molecule: vpr protein;PDBTitle: structure of human immunodeficiency virus type 1 vpr(34-51)2 peptide in dpc micelle containing aqueous solution
83 c1kzsA_
not modelled
5.2
29
PDB header: viral proteinChain: A: PDB Molecule: vpr protein;PDBTitle: structure of human immunodeficiency virus type 1 vpr(34-51)2 peptide in aqueous tfe solution
84 c6owkA_
not modelled
5.2
30
PDB header: toxinChain: A: PDB Molecule: pesticidal crystal protein cry1be, cry1k-like proteinPDBTitle: crystal structure of a bacillus thuringiensis cry1b.867 tryptic core2 variant
85 d1bw3a_
not modelled
5.2
63
Fold: Double psi beta-barrelSuperfamily: Barwin-like endoglucanasesFamily: Barwin
86 c3ofgA_
not modelled
5.1
22
PDB header: chaperoneChain: A: PDB Molecule: boca/mesd chaperone for ywtd beta-propeller-egf protein 1;PDBTitle: structured domain of caenorhabditis elegans bmy-1
87 c5v6iA_
not modelled
5.1
60
PDB header: sugar binding proteinChain: A: PDB Molecule: tmv resistance protein y3;PDBTitle: glycan binding protein y3 from mushroom coprinus comatus possesses2 anti-leukemic activity - pt derivative
88 c6cfzF_
not modelled
5.1
26
PDB header: nuclear proteinChain: F: PDB Molecule: dad1,dad1;PDBTitle: structure of the dash/dam1 complex shows its role at the yeast2 kinetochore-microtubule interface
89 c6ovbA_
not modelled
5.1
10
PDB header: toxinChain: A: PDB Molecule: active core crystal toxin protein 1d;PDBTitle: crystal structure of a bacillus thuringiensis cry1da tryptic core2 variant