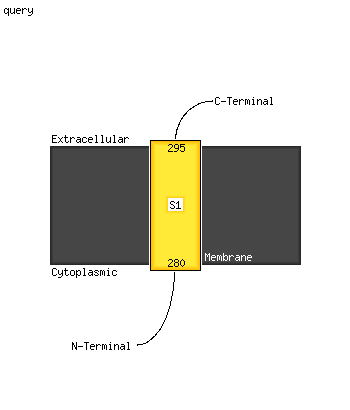
1 c3h1qB_
96.3
16
PDB header: structural proteinChain: B: PDB Molecule: ethanolamine utilization protein eutj;PDBTitle: crystal structure of ethanolamine utilization protein eutj from2 carboxydothermus hydrogenoformans
2 c4czeA_
93.4
21
PDB header: structural proteinChain: A: PDB Molecule: rod shape-determining protein mreb;PDBTitle: c. crescentus mreb, double filament, empty
3 d1dkgd2
90.5
21
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
4 c3htvA_
89.6
22
PDB header: transferaseChain: A: PDB Molecule: d-allose kinase;PDBTitle: crystal structure of d-allose kinase (np_418508.1) from escherichia2 coli k12 at 1.95 a resolution
5 d2e8aa2
89.5
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
6 c1dkgD_
88.3
21
PDB header: complex (hsp24/hsp70)Chain: D: PDB Molecule: molecular chaperone dnak;PDBTitle: crystal structure of the nucleotide exchange factor grpe2 bound to the atpase domain of the molecular chaperone dnak
7 c4kboA_
87.9
22
PDB header: signaling proteinChain: A: PDB Molecule: stress-70 protein, mitochondrial;PDBTitle: crystal structure of the human mortalin (grp75) atpase domain in the2 apo form
8 d1bupa2
87.9
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
9 c2e2pA_
87.2
14
PDB header: transferaseChain: A: PDB Molecule: hexokinase;PDBTitle: crystal structure of sulfolobus tokodaii hexokinase in2 complex with adp
10 c5hv7A_
85.0
18
PDB header: transferaseChain: A: PDB Molecule: probable sugar kinase;PDBTitle: putative sugar kinases from synechococcus elongatus pcc7942 in complex2 with d-ribulose
11 c2ap1A_
84.1
12
PDB header: transferaseChain: A: PDB Molecule: putative regulator protein;PDBTitle: crystal structure of the putative regulatory protein
12 d1jcea2
83.9
23
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
13 c1jcgA_
82.6
19
PDB header: structural proteinChain: A: PDB Molecule: rod shape-determining protein mreb;PDBTitle: mreb from thermotoga maritima, amppnp
14 d2hoea3
81.7
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK
15 c1hpmA_
81.2
23
PDB header: hydrolase (acting on acid anhydrides)Chain: A: PDB Molecule: 44k atpase fragment (n-terminal) of 7o kd heat-PDBTitle: how potassium affects the activity of the molecular2 chaperone hsc70. ii. potassium binds specifically in the3 atpase active site
16 d2zgya2
76.4
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
17 c6izrK_
76.1
22
PDB header: protein fibrilChain: K: PDB Molecule: putative plasmid segregation protein parm;PDBTitle: whole structure of a 15-stranded parm filament from clostridium2 botulinum
18 c2khoA_
76.0
21
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 70;PDBTitle: nmr-rdc / xray structure of e. coli hsp70 (dnak) chaperone (1-605)2 complexed with adp and substrate
19 d2ap1a2
75.0
7
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK
20 c3wqtB_
74.3
19
PDB header: structural genomicsChain: B: PDB Molecule: cell division protein ftsa;PDBTitle: staphylococcus aureus ftsa complexed with amppnp
21 c4rtfD_
not modelled
72.5
22
PDB header: chaperoneChain: D: PDB Molecule: chaperone protein dnak;PDBTitle: crystal structure of molecular chaperone dnak from mycobacterium2 tuberculosis h37rv
22 c6gfaA_
not modelled
71.9
16
PDB header: chaperoneChain: A: PDB Molecule: heat shock protein 105 kda;PDBTitle: structure of nucleotide binding domain of hsp110, atp and mg2+2 complexed
23 c5jygA_
not modelled
71.8
26
PDB header: structural proteinChain: A: PDB Molecule: actin-like atpase;PDBTitle: cryo-em structure of the mamk filament at 6.5 a
24 c3d2fC_
not modelled
71.0
21
PDB header: chaperoneChain: C: PDB Molecule: heat shock protein homolog sse1;PDBTitle: crystal structure of a complex of sse1p and hsp70
25 c2v7zA_
not modelled
70.6
22
PDB header: chaperoneChain: A: PDB Molecule: heat shock cognate 71 kda protein;PDBTitle: crystal structure of the 70-kda heat shock cognate protein2 from rattus norvegicus in post-atp hydrolysis state
26 c2v7yA_
not modelled
67.8
22
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein dnak;PDBTitle: crystal structure of the molecular chaperone dnak from2 geobacillus kaustophilus hta426 in post-atp hydrolysis3 state
27 c2w40C_
not modelled
67.0
11
PDB header: transferaseChain: C: PDB Molecule: glycerol kinase, putative;PDBTitle: crystal structure of plasmodium falciparum glycerol kinase2 with bound glycerol
28 c4htlA_
not modelled
65.6
10
PDB header: transferaseChain: A: PDB Molecule: beta-glucoside kinase;PDBTitle: lmo2764 protein, a putative n-acetylmannosamine kinase, from listeria2 monocytogenes
29 c3c7nB_
not modelled
64.6
23
PDB header: chaperone/chaperoneChain: B: PDB Molecule: heat shock cognate;PDBTitle: structure of the hsp110:hsc70 nucleotide exchange complex
30 d1z6ra2
not modelled
64.6
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK
31 c4ijaA_
not modelled
62.9
11
PDB header: protein bindingChain: A: PDB Molecule: xylr protein;PDBTitle: structure of s. aureus methicillin resistance factor mecr2
32 c5vm1A_
not modelled
61.7
18
PDB header: transferaseChain: A: PDB Molecule: xylulokinase;PDBTitle: crystal structure of a xyloylose kinase from brucella ovis
33 c5e84B_
not modelled
60.5
21
PDB header: chaperoneChain: B: PDB Molecule: 78 kda glucose-regulated protein;PDBTitle: atp-bound state of bip
34 c3gbtA_
not modelled
60.3
15
PDB header: transferaseChain: A: PDB Molecule: gluconate kinase;PDBTitle: crystal structure of gluconate kinase from lactobacillus acidophilus
35 c4db3A_
not modelled
58.1
16
PDB header: transferaseChain: A: PDB Molecule: n-acetyl-d-glucosamine kinase;PDBTitle: 1.95 angstrom resolution crystal structure of n-acetyl-d-glucosamine2 kinase from vibrio vulnificus.
36 c1e4gT_
not modelled
57.4
20
PDB header: bacterial cell divisionChain: T: PDB Molecule: cell division protein ftsa;PDBTitle: ftsa (atp-bound form) from thermotoga maritima
37 c3eo3B_
not modelled
57.1
22
PDB header: isomerase, transferaseChain: B: PDB Molecule: bifunctional udp-n-acetylglucosamine 2-epimerase/n-PDBTitle: crystal structure of the n-acetylmannosamine kinase domain of human2 gne protein
38 c2d0oA_
not modelled
56.4
20
PDB header: chaperoneChain: A: PDB Molecule: diol dehydratase-reactivating factor largePDBTitle: strcuture of diol dehydratase-reactivating factor complexed2 with adp and mg2+
39 c1mwmA_
not modelled
55.0
17
PDB header: structural proteinChain: A: PDB Molecule: parm;PDBTitle: parm from plasmid r1 adp form
40 c3vpzA_
not modelled
52.4
18
PDB header: transferaseChain: A: PDB Molecule: glucokinase;PDBTitle: crystal structure of glucokinase from antarctic psychrotroph at 1.69a
41 c5nckA_
not modelled
52.1
13
PDB header: transferaseChain: A: PDB Molecule: n-acetylmannosamine kinase;PDBTitle: the crystal structure of n-acetylmannosamine kinase in fusobacterium2 nucleatum
42 c3vovC_
not modelled
51.2
16
PDB header: transferaseChain: C: PDB Molecule: glucokinase;PDBTitle: crystal structure of rok hexokinase from thermus thermophilus
43 d2gupa1
not modelled
51.0
12
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK
44 c3iucC_
not modelled
49.0
17
PDB header: chaperoneChain: C: PDB Molecule: heat shock 70kda protein 5 (glucose-regulatedPDBTitle: crystal structure of the human 70kda heat shock protein 52 (bip/grp78) atpase domain in complex with adp
45 c2cgkB_
not modelled
48.7
9
PDB header: transferaseChain: B: PDB Molecule: l-rhamnulose kinase;PDBTitle: crystal structure of l-rhamnulose kinase from escherichia coli in an2 open uncomplexed conformation.
46 c1xc3A_
not modelled
48.3
13
PDB header: transferaseChain: A: PDB Molecule: putative fructokinase;PDBTitle: structure of a putative fructokinase from bacillus subtilis
47 c3mcpA_
not modelled
47.2
11
PDB header: transferaseChain: A: PDB Molecule: glucokinase;PDBTitle: crystal structure of glucokinase (bdi_1628) from parabacteroides2 distasonis atcc 8503 at 3.00 a resolution
48 c5obuA_
not modelled
44.8
19
PDB header: chaperoneChain: A: PDB Molecule: chaperone protein dnak;PDBTitle: mycoplasma genitalium dnak deletion mutant lacking sbdalpha in complex2 with amppnp.
49 d1z05a3
not modelled
44.2
16
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK
50 d3bzka5
not modelled
43.3
24
Fold: Ribonuclease H-like motifSuperfamily: Ribonuclease H-likeFamily: Tex RuvX-like domain-like
51 d1woqa1
not modelled
40.8
14
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK
52 d1pv8a_
not modelled
38.9
25
Fold: TIM beta/alpha-barrelSuperfamily: AldolaseFamily: 5-aminolaevulinate dehydratase, ALAD (porphobilinogen synthase)
53 d2aa4a1
not modelled
35.3
10
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: ROK
54 d1jcea1
not modelled
35.2
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
55 c2hoeA_
not modelled
33.7
18
PDB header: transferaseChain: A: PDB Molecule: n-acetylglucosamine kinase;PDBTitle: crystal structure of n-acetylglucosamine kinase (tm1224) from2 thermotoga maritima at 2.46 a resolution
56 d2fsja1
not modelled
32.3
19
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Ta0583-like
57 c3imoC_
not modelled
31.1
14
PDB header: unknown functionChain: C: PDB Molecule: integron cassette protein;PDBTitle: structure from the mobile metagenome of vibrio cholerae. integron2 cassette protein vch_cass14
58 d1dkgd1
not modelled
31.1
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
59 d1bdga1
not modelled
30.9
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
60 d1e4ft2
not modelled
29.3
25
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
61 c1z05A_
not modelled
28.0
13
PDB header: transcriptionChain: A: PDB Molecule: transcriptional regulator, rok family;PDBTitle: crystal structure of the rok family transcriptional regulator, homolog2 of e.coli mlc protein.
62 d1r59o1
not modelled
27.4
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Glycerol kinase
63 c3hz6A_
not modelled
26.2
17
PDB header: transferaseChain: A: PDB Molecule: xylulokinase;PDBTitle: crystal structure of xylulokinase from chromobacterium violaceum
64 c3r8eA_
not modelled
26.0
11
PDB header: transferaseChain: A: PDB Molecule: hypothetical sugar kinase;PDBTitle: crystal structure of a putative sugar kinase (chu_1875) from cytophaga2 hutchinsonii atcc 33406 at 1.65 a resolution
65 c5f7pA_
not modelled
25.4
17
PDB header: transcriptionChain: A: PDB Molecule: lmo0178 protein;PDBTitle: rok repressor lmo0178 from listeria monocytogenes
66 d1zc6a1
not modelled
25.3
15
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like
67 d2i4ra1
not modelled
25.0
10
Fold: AtpF-likeSuperfamily: AtpF-likeFamily: AtpF-like
68 d2p3ra1
not modelled
24.6
11
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Glycerol kinase
69 d1bupa1
not modelled
24.3
19
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
70 c1ig8A_
not modelled
23.6
16
PDB header: transferaseChain: A: PDB Molecule: hexokinase pii;PDBTitle: crystal structure of yeast hexokinase pii with the correct2 amino acid sequence
71 c3wxiB_
not modelled
21.9
9
PDB header: transferaseChain: B: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of trypanosoma brucei gambiense glycerol kinase2 (ligand-free form)
72 d2e8aa1
not modelled
20.3
17
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Actin/HSP70
73 c4apwH_
not modelled
19.9
11
PDB header: structural proteinChain: H: PDB Molecule: alp12;PDBTitle: alp12 filament structure
74 d1ig8a1
not modelled
18.6
16
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: Hexokinase
75 c2nlxA_
not modelled
18.3
21
PDB header: transferaseChain: A: PDB Molecule: xylulose kinase;PDBTitle: crystal structure of the apo e. coli xylulose kinase
76 c2d4wA_
not modelled
18.1
19
PDB header: transferaseChain: A: PDB Molecule: glycerol kinase;PDBTitle: crystal structure of glycerol kinase from cellulomonas sp.2 nt3060
77 c2gupA_
not modelled
17.1
9
PDB header: transferaseChain: A: PDB Molecule: rok family protein;PDBTitle: structural genomics, the crystal structure of a rok family protein2 from streptococcus pneumoniae tigr4 in complex with sucrose
78 c2i4rA_
not modelled
17.0
10
PDB header: hydrolaseChain: A: PDB Molecule: v-type atp synthase subunit f;PDBTitle: crystal structure of the v-type atp synthase subunit f from2 archaeoglobus fulgidus. nesg target gr52a.
79 c1e1cA_
not modelled
16.1
14
PDB header: isomeraseChain: A: PDB Molecule: methylmalonyl-coa mutase alpha chain;PDBTitle: methylmalonyl-coa mutase h244a mutant
80 c4c23A_
not modelled
15.5
9
PDB header: transferaseChain: A: PDB Molecule: l-fuculose kinase fuck;PDBTitle: l-fuculose kinase
81 c1xupO_
not modelled
15.3
22
PDB header: transferaseChain: O: PDB Molecule: glycerol kinase;PDBTitle: enterococcus casseliflavus glycerol kinase complexed with glycerol
82 c1v8dC_
not modelled
15.0
32
PDB header: structural genomics, unknown functionChain: C: PDB Molecule: hypothetical protein (tt1679);PDBTitle: crystal structure of the conserved hypothetical protein2 tt1679 from thermus thermophilus
83 c2fsnB_
not modelled
14.6
19
PDB header: structural proteinChain: B: PDB Molecule: hypothetical protein ta0583;PDBTitle: crystal structure of ta0583, an archaeal actin homolog, complex with2 adp
84 c1nm3B_
not modelled
14.4
18
PDB header: electron transportChain: B: PDB Molecule: protein hi0572;PDBTitle: crystal structure of heamophilus influenza hybrid-prx5
85 c3hm8D_
not modelled
12.5
12
PDB header: transferaseChain: D: PDB Molecule: hexokinase-3;PDBTitle: crystal structure of the c-terminal hexokinase domain of human hk3
86 c3ifrB_
not modelled
12.2
24
PDB header: transferaseChain: B: PDB Molecule: carbohydrate kinase, fggy;PDBTitle: the crystal structure of xylulose kinase from rhodospirillum rubrum
87 d2hiya1
not modelled
11.7
31
Fold: SP0830-likeSuperfamily: SP0830-likeFamily: SP0830-like
88 d1v8da_
not modelled
11.7
29
Fold: TTHA0583/YokD-likeSuperfamily: TTHA0583/YokD-likeFamily: Hypothetical protein TT1679
89 c2w7nA_
not modelled
11.7
20
PDB header: transcription/dnaChain: A: PDB Molecule: trfb transcriptional repressor protein;PDBTitle: crystal structure of kora bound to operator dna: insight into2 repressor cooperation in rp4 gene regulation
90 d1huxa_
not modelled
11.6
19
Fold: Ribonuclease H-like motifSuperfamily: Actin-like ATPase domainFamily: BadF/BadG/BcrA/BcrD-like
91 c1v4sA_
not modelled
11.3
15
PDB header: transferaseChain: A: PDB Molecule: glucokinase isoform 2;PDBTitle: crystal structure of human glucokinase
92 c5htxA_
not modelled
11.3
16
PDB header: transferaseChain: A: PDB Molecule: putative xylulose kinase;PDBTitle: putative sugar kinases from arabidopsis thaliana in complex with adp
93 c3js6A_
not modelled
11.3
24
PDB header: unknown functionChain: A: PDB Molecule: uncharacterized parm protein;PDBTitle: crystal structure of apo psk41 parm protein
94 c2eq5D_
not modelled
10.6
18
PDB header: isomeraseChain: D: PDB Molecule: 228aa long hypothetical hydantoin racemase;PDBTitle: crystal structure of hydantoin racemase from pyrococcus horikoshii ot3
95 c4ldnA_
not modelled
10.2
13
PDB header: transferaseChain: A: PDB Molecule: purine nucleoside phosphorylase deod-type;PDBTitle: crystal structure of a putative purine nucleoside phosphorylase from2 vibrio fischeri es114 (target nysgrc-029521)
96 c3g85A_
not modelled
10.1
11
PDB header: transcription regulatorChain: A: PDB Molecule: transcriptional regulator (laci family);PDBTitle: crystal structure of laci family transcription regulator from2 clostridium acetobutylicum
97 d1efpb_
not modelled
10.1
15
Fold: Adenine nucleotide alpha hydrolase-likeSuperfamily: Adenine nucleotide alpha hydrolases-likeFamily: ETFP subunits
98 c2yx5A_
not modelled
10.0
22
PDB header: structural genomics, unknown functionChain: A: PDB Molecule: upf0062 protein mj1593;PDBTitle: crystal structure of methanocaldococcus jannaschii purs, one of the2 subunits of formylglycinamide ribonucleotide amidotransferase in the3 purine biosynthetic pathway
99 c4j8fA_
not modelled
10.0
14
PDB header: chaperoneChain: A: PDB Molecule: heat shock 70 kda protein 1a/1b, hsc70-interacting protein;PDBTitle: crystal structure of a fusion protein containing the nbd of hsp70 and2 the middle domain of hip